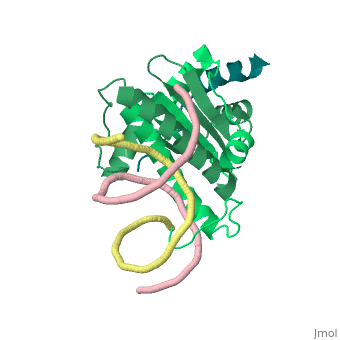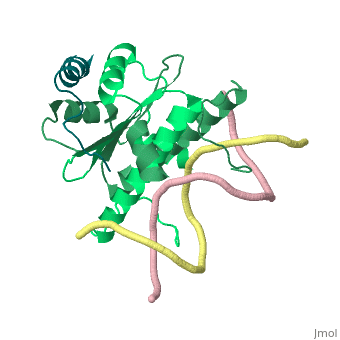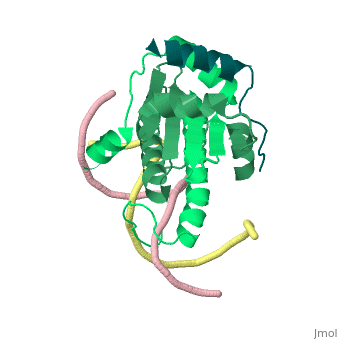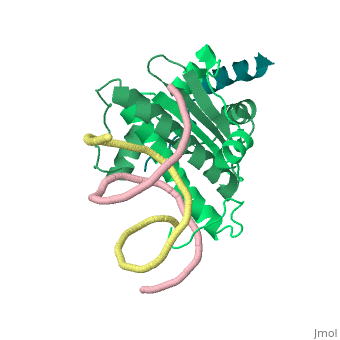Structural highlights
Function
[HDAC9_MOUSE] Devoided of intrinsic deacetylase activity, promotes the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4) by recruiting HDAC1 and HDAC3. Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Represses MEF2-dependent transcription, inhibits skeletal myogenesis and may be involved in heart development. Protects neurons from apoptosis, both by inhibiting JUN phosphorylation by MAPK10 and by repressing JUN transcription via HDAC1 recruitment to JUN promoter.[1] [2] [3] [4] [MEF2B_HUMAN] Transcriptional activator which binds specifically to the MEF2 element, 5'-YTA[AT](4)TAR-3', found in numerous muscle-specific genes. Activates transcription via this element. May be involved in muscle-specific and/or growth factor-related transcription.
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
Class II histone deacetylases (HDACs) bind myocyte enhancer factor-2 (MEF2) and repress specific gene expression in a calcium-dependent manner. Despite their significant physiological functions in muscle, immune and neuronal cells, the mechanism of recruitment of class II HDACs by MEF2 is not well understood. Here, we have characterized the complex between the MEF2-binding motif of class II HDACs and the MADS-box/MEF2S domain of MEF2B by structural and biochemical methods. The crystal structure of a HDAC9/MEF2/DNA complex reveals that HDAC9 binds to a hydrophobic groove of the MEF2 dimer. The overall binding mode is similar to that seen in the Cabin1/MEF2/DNA complex. The detailed binding interactions at the HDAC9/MEF2 interface, however, show marked differences from those at the Cabin1/MEF2 interface. Our studies suggest a general mechanism by which class II HDACs and possibly other transcriptional co-regulators are recruited by MEF2. On the other hand, the differential binding between MEF2 and its various partners may confer specific regulatory and functional properties to MEF2 in distinct cellular processes. Such specificity provides a basis for selectively disrupting a particular MEF2/co-regulator complex by mutations or small molecules.
Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2.,Han A, He J, Wu Y, Liu JO, Chen L J Mol Biol. 2005 Jan 7;345(1):91-102. PMID:15567413[5]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7354-9. Epub 2001 Jun 5. PMID:11390982 doi:http://dx.doi.org/10.1073/pnas.131198498
- ↑ Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002 Aug 23;110(4):479-88. PMID:12202037
- ↑ Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005 Mar;8(3):313-21. Epub 2005 Feb 13. PMID:15711539 doi:http://dx.doi.org/nn1408
- ↑ Morrison BE, Majdzadeh N, Zhang X, Lyles A, Bassel-Duby R, Olson EN, D'Mello SR. Neuroprotection by histone deacetylase-related protein. Mol Cell Biol. 2006 May;26(9):3550-64. PMID:16611996 doi:http://dx.doi.org/10.1128/MCB.26.9.3550-3564.2006
- ↑ Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J Mol Biol. 2005 Jan 7;345(1):91-102. PMID:15567413 doi:http://dx.doi.org/10.1016/j.jmb.2004.10.033