We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3gpd
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
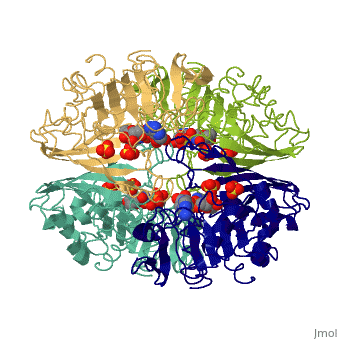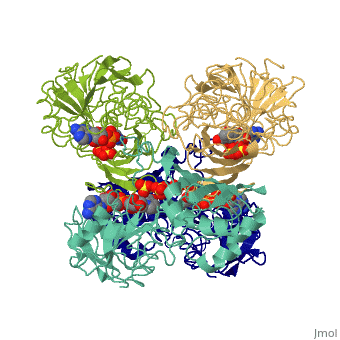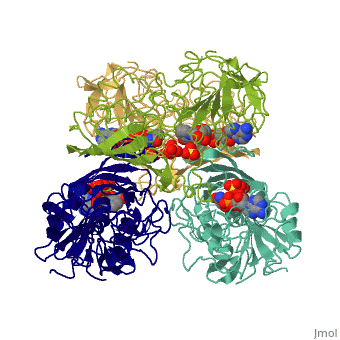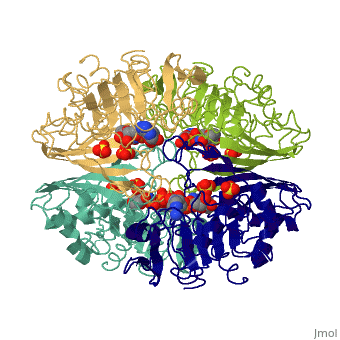
<StructureSection load='3gpd' size='340' side='right' caption='[[3gpd]], [[Resolution|resolution]] 3.50Å' scene=''> | <StructureSection load='3gpd' size='340' side='right' caption='[[3gpd]], [[Resolution|resolution]] 3.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[3gpd]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[3gpd]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Human Human]. The February 2004 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''The Glycolytic Enzymes'' by David S. Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2004_2 10.2210/rcsb_pdb/mom_2004_2]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3GPD OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3GPD FirstGlance]. <br> |
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=NAD:NICOTINAMIDE-ADENINE-DINUCLEOTIDE'>NAD</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=NAD:NICOTINAMIDE-ADENINE-DINUCLEOTIDE'>NAD</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | ||
<tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Glyceraldehyde-3-phosphate_dehydrogenase_(phosphorylating) Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating)], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.2.1.12 1.2.1.12] </span></td></tr> | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Glyceraldehyde-3-phosphate_dehydrogenase_(phosphorylating) Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating)], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.2.1.12 1.2.1.12] </span></td></tr> | ||
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3gpd FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3gpd OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3gpd RCSB], [http://www.ebi.ac.uk/pdbsum/3gpd PDBsum]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3gpd FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3gpd OCA], [http://pdbe.org/3gpd PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=3gpd RCSB], [http://www.ebi.ac.uk/pdbsum/3gpd PDBsum]</span></td></tr> |
</table> | </table> | ||
| + | == Function == | ||
| + | [[http://www.uniprot.org/uniprot/G3P_HUMAN G3P_HUMAN]] Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively. Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules (By similarity). Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation.<ref>PMID:3170585</ref> <ref>PMID:11724794</ref> <ref>PMID:23071094</ref> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 15: | Line 17: | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
| - | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3gpd ConSurf]. |
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
==See Also== | ==See Also== | ||
| + | *[[Aldehyde dehydrogenase|Aldehyde dehydrogenase]] | ||
*[[Glyceraldehyde-3-Phosphate Dehydrogenase|Glyceraldehyde-3-Phosphate Dehydrogenase]] | *[[Glyceraldehyde-3-Phosphate Dehydrogenase|Glyceraldehyde-3-Phosphate Dehydrogenase]] | ||
| + | == References == | ||
| + | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Human]] |
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: The Glycolytic Enzymes]] | [[Category: The Glycolytic Enzymes]] | ||
[[Category: Campbell, J C]] | [[Category: Campbell, J C]] | ||
[[Category: Watson, H C]] | [[Category: Watson, H C]] | ||
Revision as of 03:54, 9 February 2016
TWINNING IN CRYSTALS OF HUMAN SKELETAL MUSCLE D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE
| |||||||||||