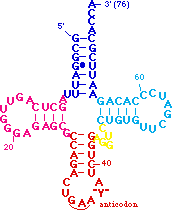We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Transfer RNA tour
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
===Domains=== | ===Domains=== | ||
| + | <scene name='72/725890/Trna_tertcolor/3'>Color the tRNA 3D structure to match the secondary structure below</scene>. | ||
[[Image:JnTphe.GIF]] | [[Image:JnTphe.GIF]] | ||
| - | <scene name='72/725890/Trna_tertcolor/3'>Color the tRNA 3D structure to match the secondary structure above</scene>. | ||
The <B><FONT COLOR="#0000ff">acceptor stem</FONT></B> includes the 5' and 3' ends of the tRNA. The 5' end is generated by RNaseP :-). The 3' end is the site which is charged with amino acids for translation. Some aminoacyl tRNA synthetases interact with both the <B><FONT COLOR="#0000ff">acceptor</FONT></B> 3' end and | The <B><FONT COLOR="#0000ff">acceptor stem</FONT></B> includes the 5' and 3' ends of the tRNA. The 5' end is generated by RNaseP :-). The 3' end is the site which is charged with amino acids for translation. Some aminoacyl tRNA synthetases interact with both the <B><FONT COLOR="#0000ff">acceptor</FONT></B> 3' end and | ||
the <B><FONT COLOR="#ff0000">anticodon</FONT></B> when charging tRNAs. Note how far the 3' end is from the <B><FONT COLOR="#ff0000">anticodon loop</FONT></B>, at bottom, by clicking here. Note also how the <B><FONT COLOR="#0000ff">acceptor stem</FONT></B> stacks onto the <B><FONT COLOR="#008aff">TpsiC stem</FONT></B> to form a continuous helix. The <B><FONT COLOR="#ff0000">anticodon stem</FONT></B> also stacks onto the junction between the <B><FONT COLOR="#aaaa00">variable loop</FONT></B> and the <B><FONT COLOR="#ff00ff">D stem</FONT></B> to form another nearly perfect helix. The <B><FONT COLOR="#008aff">TpsiC</FONT></B> and <B><FONT COLOR="#ff00ff">D loops</FONT></B> interact to bring the "cloverleaf" secondary structure in to the L-shaped tertiary structure. | the <B><FONT COLOR="#ff0000">anticodon</FONT></B> when charging tRNAs. Note how far the 3' end is from the <B><FONT COLOR="#ff0000">anticodon loop</FONT></B>, at bottom, by clicking here. Note also how the <B><FONT COLOR="#0000ff">acceptor stem</FONT></B> stacks onto the <B><FONT COLOR="#008aff">TpsiC stem</FONT></B> to form a continuous helix. The <B><FONT COLOR="#ff0000">anticodon stem</FONT></B> also stacks onto the junction between the <B><FONT COLOR="#aaaa00">variable loop</FONT></B> and the <B><FONT COLOR="#ff00ff">D stem</FONT></B> to form another nearly perfect helix. The <B><FONT COLOR="#008aff">TpsiC</FONT></B> and <B><FONT COLOR="#ff00ff">D loops</FONT></B> interact to bring the "cloverleaf" secondary structure in to the L-shaped tertiary structure. | ||
| + | ===Core Tertiary Interactions=== | ||
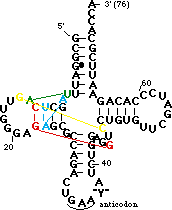
| + | Now <scene name='72/725890/Trna_tert_interact/1'> highlight the tertiary interactions</scene>. | ||
| + | Most of the backbone is shown in ribbon format, with the same color scheme as above. Several unusual base pairs, base triples and turns are highlighted and color-coded. | ||
| + | [[Image:JnTerts.GIF]] | ||
| - | < | + | The yellow residues are a parallel base pair (compared to the normal anti parallel) between <B><FONT COLOR="#ffff00">G15</FONT></B> of the <B>D-loop</B> and <B><FONT COLOR="#ffff00">C48</FONT></B> of the <B>variable loop</B>. This brings the <B>D-loop</B> and <B>variable loop</B> together. Note the sharp turn in the backbone between <B><FONT COLOR="#ffff00">C48</FONT></B> and <B><FONT COLOR="#ffff00">C49</FONT></B> caused by the parallel pair.</P> |
| - | + | ||
| - | + | The green residues are a reverse Hoogsteen pair between <B><FONT COLOR="#00bf00">U8</FONT></B>and <B><FONT COLOR="#00bf00">A14</FONT></B>. This pairing is important for positioning of the <B>D stem</B> relative to the stacked <B>T</B> and <B>acceptor</B> stems.</P> | |
| + | |||
| + | The cyan residues are a base triple in which <B><FONT COLOR="#008aff">A9</FONT></B>H-bonds in the major groove to <B><FONT COLOR="#008aff">A23</FONT></B> (which is paired with <B><FONT COLOR="#008aff">U12</FONT></B>). It stabilizes a sharp turn between bases 9 and 10.</P> | ||
| + | |||
| + | The red residues are a base triple in which <B><FONT COLOR="#ff0000">7-methyl-G46</FONT></B> from the <B>variable loop</B> H-bonds to the <B><FONT COLOR="#ff0000">G22-C13</FONT></B> base pair of the <B>D stem</B>. This helps dock the <B>variable loop</B> onto the <B>D-stem</B>.</P> | ||
| - | You can <scene name='72/725869/Zoom_pairs/1'>look at just four of the base pairs.</scene>.You are looking into the major groove and the colors of the base pairs alternate. You can also <scene name='72/725869/Zoom_pairs_only/1'>look at just the bases</scene>. | ||
| - | Each base pair stacks on the next similarly, as shown from <scene name='72/725869/Zoom_pairs_top/1'>this top view</scene>. This is the <scene name='72/725869/Zoom_pairs_only_top/1'>same top view of just the bases</scene>. | ||
| - | B-DNA stacks similarly, but compare this with Z-DNA, which behaves much differently. Essentially all helical RNA is in A form, but DNA can also be found in A form under certain conditions (particularly in RNA-DNA hybrids). The 2'-OH of ribose favors the C3'-''endo'' sugar pucker necessary for A-form geometry. The O2' is easily seen as white spheres in <scene name='72/725869/Rna_space_filling_o2prime/1'>this space fill view</scene>. | ||
| - | You can compare it with the DNA forms by looking at this [http://proteopedia.org/wiki/images/d/d3/JnABZ3d.gif 3D red-blue stereo picture of A, B, and Z DNA] | ||
| - | </StructureSection> | ||
| - | ==See Also== | ||
| - | * [[Z-DNA model tour]] and [[Z-DNA]] | ||
| - | * [[B-DNA tour]] | ||
| - | * A more general overview will be found at [[DNA]]. | ||
| - | * [[Forms of DNA]] shows a side-by-side comparison of A, B, and Z forms of DNA. | ||
| - | * An interactive tutorial on [http://dna.molviz.org DNA Structure], ''disponible también en español'' and eight other languages. | ||
== References == | == References == | ||
Revision as of 16:57, 25 February 2016
A-form RNA
| |||||||||||