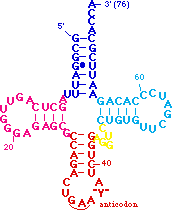We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Transfer RNA tour
From Proteopedia
(Difference between revisions)
m |
m |
||
| Line 15: | Line 15: | ||
[[Image:JnTerts.GIF]] | [[Image:JnTerts.GIF]] | ||
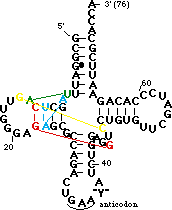
| - | <scene name='72/725890/Trna_tert_interact/ | + | <scene name='72/725890/Trna_tert_interact/8'>You can zoom in for a closer look</scene>. The yellow residues are a parallel base pair (compared to the normal anti parallel) between <B><FONT COLOR="#aaaa00">G15</FONT></B> of the <B>D-loop</B> and <B><FONT COLOR="#aaaa00">C48</FONT></B> of the <B>variable loop</B>. This brings the <B>D-loop</B> and <B>variable loop</B> together. Note the sharp turn in the backbone between <B><FONT COLOR="#aaaa00">C48</FONT></B> and <B><FONT COLOR="#aaaa00">C49</FONT></B> caused by the parallel pair. |
The green residues are a reverse Hoogsteen pair between <B><FONT COLOR="#00bf00">U8</FONT></B> and <B><FONT COLOR="#00bf00">A14</FONT></B>. This pairing is important for positioning of the <B>D stem</B> relative to the stacked <B>T</B> and <B>acceptor</B> stems. | The green residues are a reverse Hoogsteen pair between <B><FONT COLOR="#00bf00">U8</FONT></B> and <B><FONT COLOR="#00bf00">A14</FONT></B>. This pairing is important for positioning of the <B>D stem</B> relative to the stacked <B>T</B> and <B>acceptor</B> stems. | ||
Revision as of 16:38, 3 March 2016
tRNA
| |||||||||||
See Also
- Z-DNA model tour and Z-DNA
- B-DNA tour
- A-RNA tour
- A more general overview will be found at DNA.
- Forms of DNA shows a side-by-side comparison of A, B, and Z forms of DNA.
- An interactive tutorial on DNA Structure, disponible también en español and eight other languages.
References
JSmol in Proteopedia [2] or to the article describing Jmol [3] to the rescue.
- ↑ Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796-806. PMID:790568
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644