We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
BamHI
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
==Active Sites and Catalytic Mechanism to Specific DNA== | ==Active Sites and Catalytic Mechanism to Specific DNA== | ||
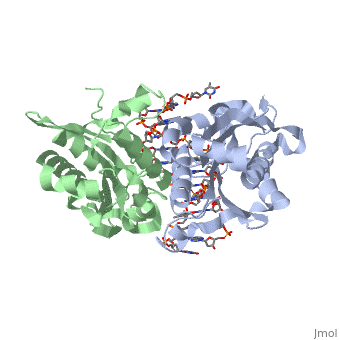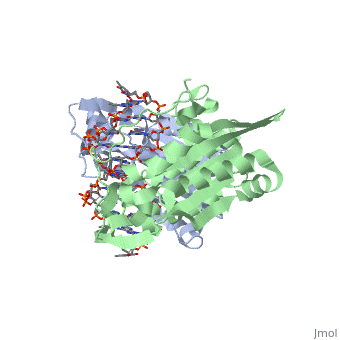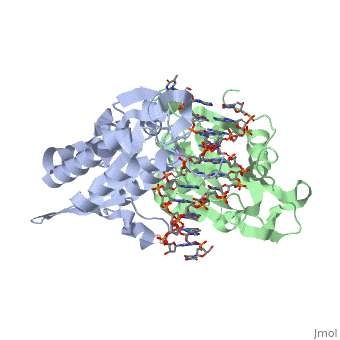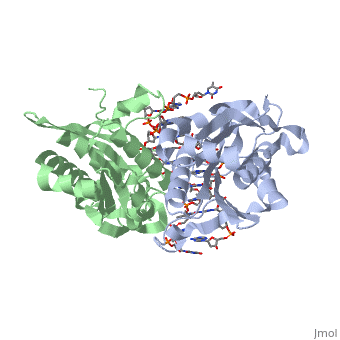
| - | Type II restriction enzymes require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA phosphodiesters, leaving free 5’ phosphate and 3’ hydroxyl groups. The reaction is considered to proceed by an inline displacement of the 3’ leaving group, in which an activated water molecule acts as the attacking nucleophile. The <scene name='47/472560/Active_site/2'>active site</scene> for ''Bam''HI are residues Asp94, Glu111, and Glu113. These can be spatially aligned with residues in EcoRI, EcoRV and PvuII. Several mechanisms have been proposed for the way these enzymes might activate a water molecule for nucleophilic attack. It is suggested there is a general base mechanism in which the acidic residues Asp94 and Glu111 coordinate Mg2+ at the active site while Glu113 acts as a general base to deprotonate the attacking water molecule. A separate mechanism has been suggested in which one of the <scene name='47/472560/Active_site_water/1'>three water molecules | + | Type II restriction enzymes require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA phosphodiesters, leaving free 5’ phosphate and 3’ hydroxyl groups. The reaction is considered to proceed by an inline displacement of the 3’ leaving group, in which an activated water molecule acts as the attacking nucleophile. The <scene name='47/472560/Active_site/2'>active site</scene> for ''Bam''HI are residues Asp94, Glu111, and Glu113. These can be spatially aligned with residues in EcoRI, EcoRV and PvuII. Several mechanisms have been proposed for the way these enzymes might activate a water molecule for nucleophilic attack. It is suggested there is a general base mechanism in which the acidic residues Asp94 and Glu111 coordinate Mg2+ at the active site while Glu113 acts as a general base to deprotonate the attacking water molecule. A separate mechanism has been suggested in which one of the <scene name='47/472560/Active_site_water/1'>three water molecules</scene> located centrally at the active site within hydrogen-bonding distances from the carboxylate groups of Glu111 and Glu113 acts as a nucleophile and attacks the phosphate group of DNA.<ref name="Newman">Newman, Matthew, Structure of ''Bam''HI Endonuclease Bound to DNA: Partial Folding and Unfolding on DNA Binding., ''Science.'' 1995 Aug;4(269):656-63. Print.</ref> |
==Active Sites and Catalytic Mechanism for Binding to Nonspecific DNA== | ==Active Sites and Catalytic Mechanism for Binding to Nonspecific DNA== | ||
Revision as of 22:14, 17 March 2016
| |||||||||||
3D structures of BamHI
Updated on 17-March-2016
1bhm, 3bam, 2bam, 1esg – BaBAM + DNA – Bacillus amyloliquefaciens
1bam – BaBAM
References
- ↑ 1.0 1.1 Viadiu H, Aggarwal AK, Structure of BamHI bound to nonspecific DNA: a model for DNA sliding., Mol Cell. 2000 May;5(5):889-95. Print.
- ↑ Voet, Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.
- ↑ 3.0 3.1 PDBe., European Molecular Biology Laboratory. 2011. Webpage.
- ↑ Newman, Matthew, Structure of BamHI Endonuclease Bound to DNA: Partial Folding and Unfolding on DNA Binding., Science. 1995 Aug;4(269):656-63. Print.
Proteopedia Page Contributors and Editors (what is this?)
Shane Michael Evans, Michal Harel, Alexander Berchansky, Ann Taylor