Glycerol-3-Phosphate Dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
[[Image:FINAL.png|thumb|Glycerol 3-Phosphate Dehydrogenase]] | [[Image:FINAL.png|thumb|Glycerol 3-Phosphate Dehydrogenase]] | ||
| - | + | {{Clear}} | |
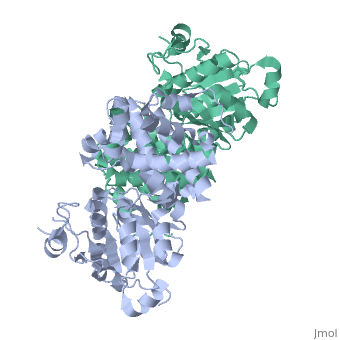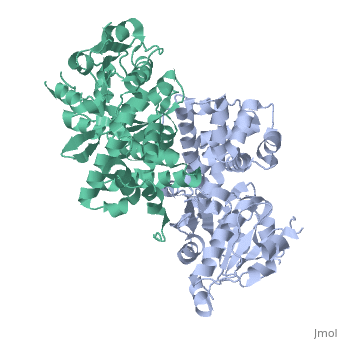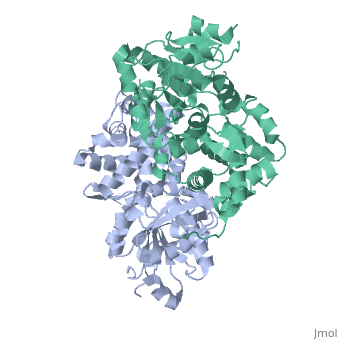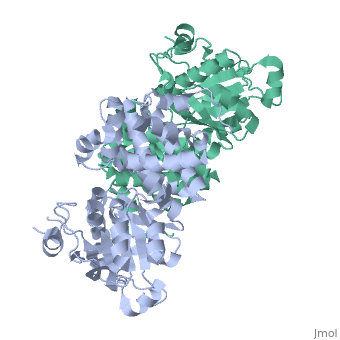
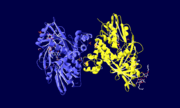
GlpD is a dimer that consists of two subunits; α and β. The GlpD structure also contains seven ligands; 1,3-Dihydroxyacetonephosphate (13P), β-Octylglucoside (βOG), 1,2-Ethanediol (EDO), Flavin-Adenine Dinucleotide (FAD), Imidazole (IMD), PO4 (Phosphate Ion) and N-(Tris(Hydroxymethyl)methyl)-3-Aminopropanesulfonic Acid (T3A). The active sites on GlpD are the Cap-Domain, FAD- Domain and a ubiquinone substrate analogue, menadione (MD). | GlpD is a dimer that consists of two subunits; α and β. The GlpD structure also contains seven ligands; 1,3-Dihydroxyacetonephosphate (13P), β-Octylglucoside (βOG), 1,2-Ethanediol (EDO), Flavin-Adenine Dinucleotide (FAD), Imidazole (IMD), PO4 (Phosphate Ion) and N-(Tris(Hydroxymethyl)methyl)-3-Aminopropanesulfonic Acid (T3A). The active sites on GlpD are the Cap-Domain, FAD- Domain and a ubiquinone substrate analogue, menadione (MD). | ||
Revision as of 09:35, 20 March 2016
| |||||||||||
3D structures of glycerol-3-phosphate dehydrogenase
Updated on 20-March-2016
References
- ↑ Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3280-5. Epub 2008 Feb 22. PMID:18296637
- ↑ Yeh JI, Charrier V, Paulo J, Hou L, Darbon E, Claiborne A, Hol WG, Deutscher J. Structures of enterococcal glycerol kinase in the absence and presence of glycerol: correlation of conformation to substrate binding and a mechanism of activation by phosphorylation. Biochemistry. 2004 Jan 20;43(2):362-73. PMID:14717590 doi:10.1021/bi034258o
Proteopedia Page Contributors and Editors (what is this?)
Indu Toora, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell, Andrew Rebeyka, Jaime Prilusky