We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1176
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| + | ==Introduction== | ||
| + | |||
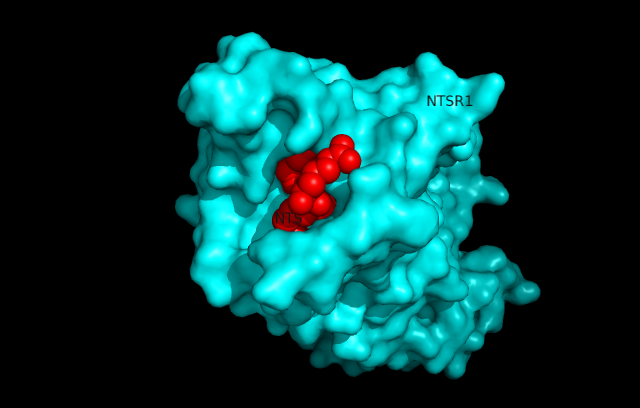
| + | Neurotensin receptor 1 (NTSR1) is a G-protein coupled receptor (GPCR) that binds to the 13 amino acid peptide, neurotensin. Studies determining the structure of NTSR1 crystallized the GPCR bound with the C-terminus of its tridecapeptide ligand, <scene name='72/721548/Neurotensin/3'>NTS(8-13)</scene> because it has a higher potency and efficacy than its full-length counterpart. NTSR1 is a class A GPCR, and like all G-proteins, consists of an extracellular binding domain along with 7 transmembrane helices. Along with the ligand binding pocket at the top of the protein, NTSR1 also contains an allosteric Na+ ion binding pocket underneath. NTS binds to NTSR1, leading to a conformational change of the protein and modulation of second messengers. NTS has been shown to have a variety of biological activities including a role in the leptin signalling pathways, tumor growth, and dopamine regulation. The majority of effects of NTS are mediated through NTSR1. Research of the structure of NTSR1 has focused on the differences between its active and active-like states. | ||
| + | [[Image:surfaceprotein.png]] | ||
== Structure == | == Structure == | ||
===Ligand Binding Pocket=== | ===Ligand Binding Pocket=== | ||
Revision as of 02:04, 30 March 2016
| This Sandbox is Reserved from Jan 11 through August 12, 2016 for use in the course CH462 Central Metabolism taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1160 through Sandbox Reserved 1184. |
To get started:
More help: Help:Editing |
Rattus norevegicus NTSR1
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644