We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1167
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Function == | == Function == | ||
| - | The | + | The glucagon receptor plays an important role in glucose homeostasis. During times of fasting (or low blood sugar) the pancreas dispatches glucagon to activate the GCGR in the liver. The [http://www.nature.com/nature/journal/v499/n7459/fig_tab/nature12393_F5.html binding of glucagon] stimulates [https://en.wikipedia.org/wiki/Gluconeogenesis gluconeogenesis], through [https://en.wikipedia.org/wiki/Adenylyl_cyclase adenylate cyclase] that initiates [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A] (PKA) activity<ref>PMID:25674162</ref>. This pathway synthesizes glucose, elevating blood sugar levels. |
| Line 16: | Line 16: | ||
=== Class B vs. Class A === | === Class B vs. Class A === | ||
| - | + | In contrast to class A glucagon receptors which have a [https://en.wikipedia.org/wiki/Proline proline] kink, in all [https://en.wikipedia.org/wiki/Secretin_receptor_family secretin-like class B glucagon receptors] there is a [https://en.wikipedia.org/wiki/Glycine Glycine] at position 393 in Helix VII which allows for a <scene name='72/721537/Gly_393_helical_bend/1'>helical bend</scene>. This glycine helical bend is fully [https://en.wikipedia.org/wiki/Conserved_sequence conserved] in all secretin-like class B receptors and is an important part of the FQGxxVxxYCF [https://en.wikipedia.org/wiki/Sequence_motif motif]. | |
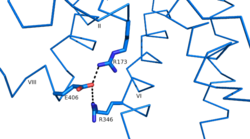
| - | Another important structural component found in all secretin-like class B receptors are the two conserved [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] found between [https://en.wikipedia.org/wiki/Arginine Arg] 346 and [https://en.wikipedia.org/wiki/Glutamic_acid Glu] 406 and Arg 173 and Glu 406 [[Image:Salt Bridge Interactions.png | 250 px|right|thumb|Glu 406 Salt Bridges]]. | + | Another important structural component found in all secretin-like class B receptors are the two conserved [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] found between [https://en.wikipedia.org/wiki/Arginine Arg] 346 and [https://en.wikipedia.org/wiki/Glutamic_acid Glu] 406 and Arg 173 and Glu 406 [[Image:Salt Bridge Interactions.png | 250 px|right|thumb|Glu 406 Salt Bridges]]. These salt bridges appear to be distinct feature of class B receptors only, however, because there is no conservation of these residues seen in class A receptors. |
| - | + | A conserved [https://en.wikipedia.org/wiki/Disulfide disulfide bond] can be found in 7tm region at <scene name='72/721538/7tm_disulfide_bond/2'>Cys224-Cys294</scene> which helps to stabilize the 7tm fold. This bond is conserved among both class A and class B receptors. Additionally, the ECD region of Class B GCPRs is defined by three conserved disulfide bonds. These bonds occur at <scene name='72/721538/Ecd_disulfide_bond_1/4'>Cys62-Cys104</scene>, <scene name='72/721538/Ecd_disulfide_bond_2/1'>Cys46-Cys71</scene>, and <scene name='72/721538/Ecd_disulfide_bond_3/1'>Cys85-Cys126</scene>. | |
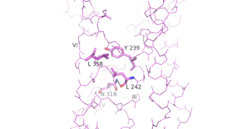
| - | As a part of the interface stabilization between helices VI, V, and III, a Class B-specific [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] occurs between [https://en.wikipedia.org/wiki/Asparagine | + | As a part of the interface stabilization between helices VI, V, and III, a Class B-specific [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] occurs between [https://en.wikipedia.org/wiki/Asparagine Asn] 318 of Helix V and [https://en.wikipedia.org/wiki/Leucine Leu] 242 of Helix III. [[Image:Conserved Class B Residues.png | 250 px|left|thumb|Helical Interactions VI-V-III]] |
=== GCGR-Specific Traits === | === GCGR-Specific Traits === | ||
| Line 30: | Line 30: | ||
==== Intracellular Helix VIII ==== | ==== Intracellular Helix VIII ==== | ||
| - | The GCGR also contains an intracellular Helix VIII that is comprised of roughly 20 amino acids at the C-terminal end. This helix tilts approximately 25 degrees away from the membrane - the corresponding position in | + | The GCGR also contains an intracellular Helix VIII that is comprised of roughly 20 amino acids at the C-terminal end. This helix tilts approximately 25 degrees away from the membrane - the corresponding position in class A receptors are turned toward the membrane<ref>PMID:23863937</ref>. Although researchers are not entirely sure of its function, this helix is completely conserved in class B structures. |
==== Binding Pocket ==== | ==== Binding Pocket ==== | ||
| - | The | + | The class B GPCR has the widest and longest binding pocket. The distance between the EC tips of Helicies II and VI as well as between the tips of Helicies III and VII are some of the largest among the GPCRs<ref>PMID:23863937</ref>. As a result, the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3820480/bin/nihms495648f2.jpg binding cavity] of GCGR is located deeper inside the molecule. |
====Other Unique Structural Features ==== | ====Other Unique Structural Features ==== | ||
| - | An important interface stabilization interaction between | + | An important interface stabilization interaction between Helices I and VII occurs between [https://en.wikipedia.org/wiki/Serine Ser] 152 of Helix I and Ser 390 of Helix VII. Due to their close proximity to one another, they form an important <scene name='72/721537/Ser-ser_hydrogen_bond/2'>hydrogen bond</scene> which stabilizes the structure of GCGR. |
== Glucagon Binding == | == Glucagon Binding == | ||
| - | Research has shown that | + | Research has shown that class B GCPRs exist in either an [http://www.nature.com/ncomms/2015/150731/ncomms8859/fig_tab/ncomms8859_F3.html open or closed conformation]. To transition between states, the ECD rotates and moves down towards the 7tm domain. The stalk region of Helix I helps to facilitate this motion of the ECD. |
In its open state, the ECD and the stalk region of Helix 1 are almost perpendicular to the membrane surface. In the case of GCGR, this open confirmation is stabilized by glucagon binding. In the absence of glucagon, however, the GCGR adopts a closed conformation in which all three of the extracellular loops of the 7tm (ECL1, ECL2, and ECL3) can interact with the ECD. In this closed state, the ECD covers the extracellular surface of the 7tm. | In its open state, the ECD and the stalk region of Helix 1 are almost perpendicular to the membrane surface. In the case of GCGR, this open confirmation is stabilized by glucagon binding. In the absence of glucagon, however, the GCGR adopts a closed conformation in which all three of the extracellular loops of the 7tm (ECL1, ECL2, and ECL3) can interact with the ECD. In this closed state, the ECD covers the extracellular surface of the 7tm. | ||
| - | This transition mechanism is consistent with the "two-domain" binding mechanism of | + | This transition mechanism is consistent with the "two-domain" binding mechanism of class B GCPRs in which (1) the C-terminus of the ligand first binds to the ECD allowing (2) the N-terminus of the ligand to interact with the 7tm and activate the protein. |
== Clinical Relevance == | == Clinical Relevance == | ||
| - | Because of GCGR's role in glucose homeostasis, it is a potential drug target for [https://en.wikipedia.org/wiki/Diabetes_mellitus_type_2 Type 2 diabetes]. Specifically, molecules that antagonize the glucagon receptor may be able to lower blood sugar levels. Studies have shown that two antibodies, mAb1 and mAb23, target the ECD domain of the GCGR | + | Because of GCGR's role in glucose homeostasis, it is a potential drug target for [https://en.wikipedia.org/wiki/Diabetes_mellitus_type_2 Type 2 diabetes]. Specifically, molecules that antagonize the glucagon receptor may be able to lower blood sugar levels. Studies have shown that two antibodies, mAb1 and mAb23, target the ECD domain of the GCGR interrupting glucagon binding<ref>PMID:22908259</ref>. By disrupting the normal interactions between the ECD and the 7tm domains, these antibiotics inhibit the receptor's function and help to lower blood glucose level. Additional research has shown that another antibody, mAb7, inhibits GCGR allosterically<ref>PMID:24189067</ref>. Through binding to a site outside of the binding pocket, mAb7 inhibits the receptor without interacting with essential glucagon binding residues. |
</StructureSection> | </StructureSection> | ||
Revision as of 16:41, 30 March 2016
| This Sandbox is Reserved from Jan 11 through August 12, 2016 for use in the course CH462 Central Metabolism taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1160 through Sandbox Reserved 1184. |
To get started:
More help: Help:Editing |
Class B Human Glucagon G-Protein Coupled Receptor
References
- ↑ Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859
- ↑ Lotfy M, Kalasz H, Szalai G, Singh J, Adeghate E. Recent Progress in the Use of Glucagon and Glucagon Receptor Antago-nists in the Treatment of Diabetes Mellitus. Open Med Chem J. 2014 Dec 31;8:28-35. doi: 10.2174/1874104501408010028., eCollection 2014. PMID:25674162 doi:http://dx.doi.org/10.2174/1874104501408010028
- ↑ Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, Clarke HJ, Wong T, Chiang V, Luis E, Estevez A, Rondon J, Zhang Y, Hotzel I, Allan BB. Molecular basis for negative regulation of the glucagon receptor. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14393-8. Epub 2012 Aug 20. PMID:22908259 doi:http://dx.doi.org/10.1073/pnas.1206734109
- ↑ Mukund S, Shang Y, Clarke HJ, Madjidi A, Corn JE, Kates L, Kolumam G, Chiang V, Luis E, Murray J, Zhang Y, Hotzel I, Koth CM, Allan BB. Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor. J Biol Chem. 2013 Nov 4. PMID:24189067 doi:http://dx.doi.org/10.1074/jbc.M113.496984