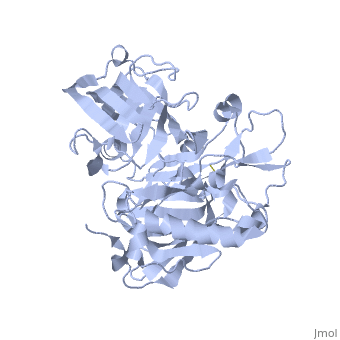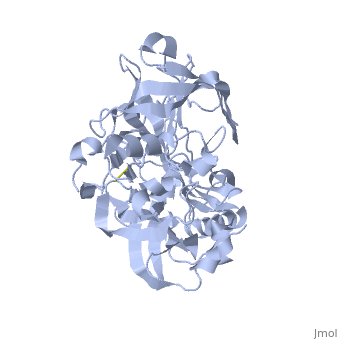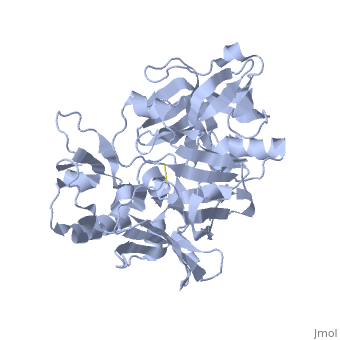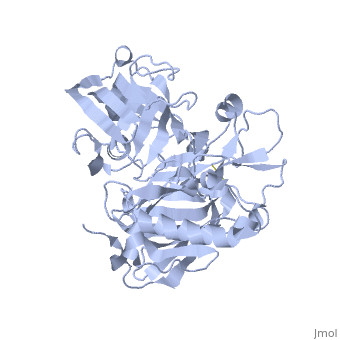Beta Secretase (BACE1) 1SGZ
From Proteopedia
(New page: Beta-secretase, also known as '''BACE''' or '''Memapsin 2''', is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellula...) |
|||
| Line 6: | Line 6: | ||
== Structure == | == Structure == | ||
| - | <StructureSection load=' | + | <StructureSection load='1SGZ' size='340' side='right' caption='Human Beta-secretase (PDB 1SGZ)' scene=''> |
BACE-1 is a type-1 integral membrane glycoprotein that is affected by the positions on the outer domains of amino acids. Cleavage of soluble substrates is possible in external environment due to the enzyme’s high values of Km failing to observe in the internal environment. BACE-1 prefers the cleavage of longer substrates over smaller peptide based substrates due to approximate acidic pH of 4.5 resulting from the cleavage. The acidic and polar environment of the enzyme allow a wide variety of substrates to bind to the membrane. The two catalytic aspartate residues that reside in the active site of BACE-1 are Asp 32 and Asp 228. The active site of BACE-1 has a flexible antiparallel -hairpin, also known as a flap, which is found in the residues of 67 to 77, that controls the accessibility of the substrate to the active site, and the flap also has to set the substrate in the correct orientation for catalysis. In an open conformation, the active site allows the substrate to enter without any difficulty and regulates its activity. In an inactive form, the binding affinity to the inhibitor is decreased, which then lowers the substrate binding and enzymatic activity. For the closed conformation, the flap with the inhibitor bound is firmly secured in place. | BACE-1 is a type-1 integral membrane glycoprotein that is affected by the positions on the outer domains of amino acids. Cleavage of soluble substrates is possible in external environment due to the enzyme’s high values of Km failing to observe in the internal environment. BACE-1 prefers the cleavage of longer substrates over smaller peptide based substrates due to approximate acidic pH of 4.5 resulting from the cleavage. The acidic and polar environment of the enzyme allow a wide variety of substrates to bind to the membrane. The two catalytic aspartate residues that reside in the active site of BACE-1 are Asp 32 and Asp 228. The active site of BACE-1 has a flexible antiparallel -hairpin, also known as a flap, which is found in the residues of 67 to 77, that controls the accessibility of the substrate to the active site, and the flap also has to set the substrate in the correct orientation for catalysis. In an open conformation, the active site allows the substrate to enter without any difficulty and regulates its activity. In an inactive form, the binding affinity to the inhibitor is decreased, which then lowers the substrate binding and enzymatic activity. For the closed conformation, the flap with the inhibitor bound is firmly secured in place. | ||
| Line 29: | Line 29: | ||
In a general reaction, APP is initially cleaved by alpha-secretase in a certain position every time and then by gamma-secretase. Neurons may benefits from the released fragments produced by the alpha, gamma-secretase cleavage of APP. However, when beta-secretase enters the equation the cleavage of APP changes positions. Once gamma-secretase has officially completed its cleavage, amyloid-beta is released into the extracellular membrane space. Amyloid-beta is most commonly found to aggregate in this area and form plaques that are extremely harmful to the human body. It is thought that this plaque build up is the initial cause of Alzheimer’s Disease. | In a general reaction, APP is initially cleaved by alpha-secretase in a certain position every time and then by gamma-secretase. Neurons may benefits from the released fragments produced by the alpha, gamma-secretase cleavage of APP. However, when beta-secretase enters the equation the cleavage of APP changes positions. Once gamma-secretase has officially completed its cleavage, amyloid-beta is released into the extracellular membrane space. Amyloid-beta is most commonly found to aggregate in this area and form plaques that are extremely harmful to the human body. It is thought that this plaque build up is the initial cause of Alzheimer’s Disease. | ||
| - | == Associated | + | == Associated Health Conditions == |
---- | ---- | ||
| - | == Alzheimer's Disease == | + | === Alzheimer's Disease === |
| - | == Down Syndrome == | + | === Down Syndrome === |
== Inhibitors == | == Inhibitors == | ||
Revision as of 12:55, 5 April 2016
Beta-secretase, also known as BACE or Memapsin 2, is encoded by the gene BACE1. Beta-secretase is a proteolytic, transmembrane enzyme with two active sites on the extracellular region. It is associated with processing amyloid precursor protein (APP), which is an integral membrane protein. A malfunction in the processing of APP results in the formation of the peptide amyloid-beta. Amyloid-beta is a neurotoxic peptide segment that aggregates into plaques. These plaques are the primary component of plaques found in individuals with Alzheimer’s disease. Other biological associations of this enzyme include modulating myelination in the central and peripheral nervous systems.
Enzyme Class
Beta-secretase is an enzyme that is classified as a class 3 enzyme, which are hydrolase enzymes. It acts on breaking peptide bonds and therefore is also considered a peptidase and belongs to the subclass of aspartic acid endopeptidases.
Structure
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644