Sandbox Reserved 428
From Proteopedia
(Difference between revisions)
| Line 39: | Line 39: | ||
==Binding Interactions== | ==Binding Interactions== | ||
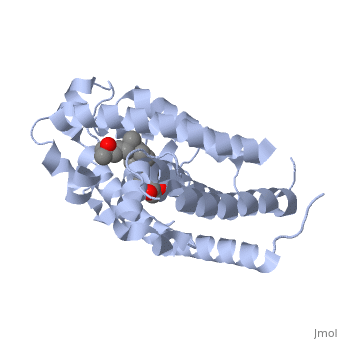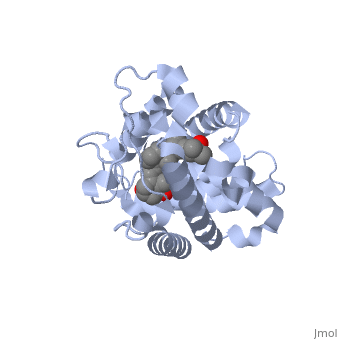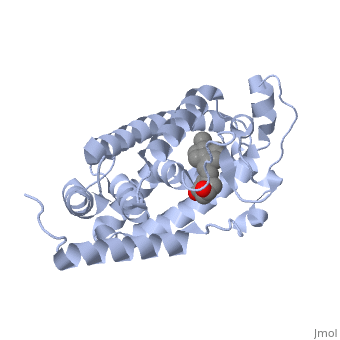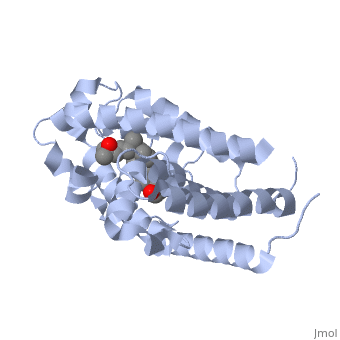
| - | Protein 1db1 is found to complex with 1,25 Dihydroxy <scene name='48/483885/Vitamin_d3/1'>Vitamin D3</scene>. This molecule has three notable alcohol groups shown in red. Oxygen is electronegative, giving alcohols the ability to participate in hydrogen bonding with the protein. Vitamin D3 has a large number of relations with the residues on the protein chain. First in the sequence are <scene name='48/483885/Residues_140-151/2'>residues 140-151</scene>. Tyr143, shown in blue, is the closest to the ligand at 2.83 angstroms. This is sightly large but there is still the possibility of hydrogen bonding. Tyr147 in green and Phe150 in black are also known to have interactions with Vitamin D3 they are farther away and therefore less significant. Next down the peptide chain are <scene name='48/483885/Residues_235-240/1'>residues 235-240</scene>. Ser237, shown in green, has significant interactions with vitamin D3 this can be seen by its short distance 2.78 angstroms. Only 40 residues away, more hydrogen bonding is occurring.<scene name='48/483885/Residues_270-280/1'>Arg274 and Ser278</scene> form bonds <scene name='48/483885/Residues_304-306/1'>His305</scene> | + | Protein 1db1 is found to complex with 1,25 Dihydroxy <scene name='48/483885/Vitamin_d3/1'>Vitamin D3</scene>. This molecule has three notable alcohol groups shown in red. Oxygen is electronegative, giving alcohols the ability to participate in hydrogen bonding with the protein. Vitamin D3 has a large number of relations with the residues on the protein chain. First in the sequence are <scene name='48/483885/Residues_140-151/2'>residues 140-151</scene>. Tyr143, shown in blue, is the closest to the ligand at 2.83 angstroms. This is sightly large but there is still the possibility of hydrogen bonding. Tyr147 in green and Phe150 in black are also known to have interactions with Vitamin D3 they are farther away and therefore less significant. Next down the peptide chain are <scene name='48/483885/Residues_235-240/1'>residues 235-240</scene>. Ser237, shown in green, has significant interactions with vitamin D3 this can be seen by its short distance 2.78 angstroms. Only 40 residues away, more hydrogen bonding is occurring.<scene name='48/483885/Residues_270-280/1'>Arg274 and Ser278</scene> form bonds with the same oxygen atoms as Tyr143 and Ser237 respectively <scene name='48/483885/Residues_304-306/1'>His305</scene> |
==Additional Features== | ==Additional Features== | ||
Revision as of 20:41, 6 April 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Vitamin D receptor/vitamin D (1db1)[1]
by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole
Student Projects for UMass Chemistry 423 Spring 2016
| |||||||||||