Sandbox Reserved 428
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
==Introduction== | ==Introduction== | ||
<br> | <br> | ||
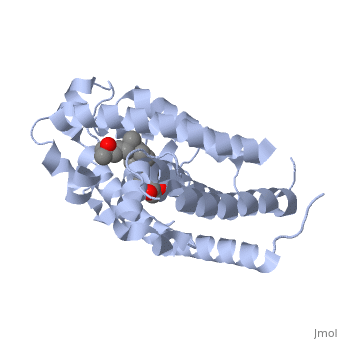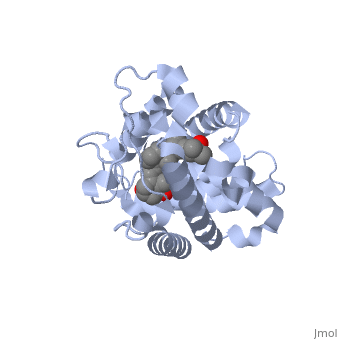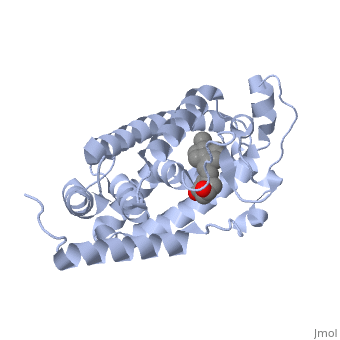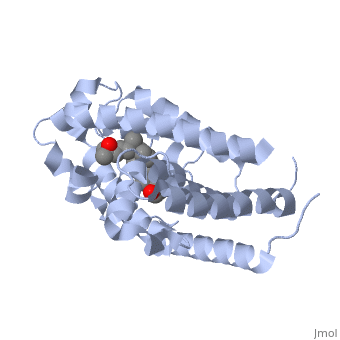
| - | The vitamin D receptor (VDR) is a ligand-dependent transcriptional regulator with two alpha helices shown in this <scene name='48/483885/Color1/6'>colored scence</scene> showing the flow of the strands starting at the blue amino end flowing to the red carboxy end. VDR belongs to the superfamily of nuclear receptors which control homeostasis, cell differentiation and growth, and many physiological processes. All proteins that belong to the nuclear receptor superfamily have a variable N-terminus region (A/B region), a hinge region that is flexible (D region), a conserved DNA-binding region (DBD, C region), and a moderately conserved ligand-binding region (LBD, E/F region). In the case of VDR, the A/B region is very short so it does not have any AF-1 function and the ligand binding region has a dimerization interface and a transcriptional activation domain that is ligand-dependent (AF-2).[1] | + | The vitamin D receptor (VDR) is a ligand-dependent transcriptional regulator with two alpha helices shown in this <scene name='48/483885/Color1/6'>colored scence</scene> showing the flow of the strands starting at the blue amino end flowing to the red carboxy end. VDR belongs to the superfamily of nuclear receptors which control homeostasis, cell differentiation and growth, and many physiological processes. All proteins that belong to the nuclear receptor superfamily have a variable N-terminus region (A/B region), a hinge region that is flexible (D region), a conserved DNA-binding region (DBD, C region), and a moderately conserved ligand-binding region (LBD, E/F region). In the case of VDR, the A/B region is very short so it does not have any AF-1 function and the ligand binding region has a dimerization interface and a transcriptional activation domain that is ligand-dependent (AF-2).[1] <br> |
The VDR has both an active and suppressed form. The activation or suppression function is caused by the binding of the DR3 response element as a heterodimer with the retinoid X receptor of the target genes. Due to the interactions with the basal transcriptional machinery and transcriptional cofactors, transcription is either activated or suppressed. When VDR is in its active form it regulates both phosphate and calcium metabolism, has immunosuppressive effects, and induces cell differentiation. When there are defects in the VDR that effect its metabolism it can lead to diseases such as severe rickets, secondary hyperparathyroidism, and hypocalcemia. Though defects in VDR can cause many diseases, fully functioning VDR can be used as treatment for disease such as cancer, autoimmune disease, psoriasis, osteoporosis, and renal osteodystrophy.[1] | The VDR has both an active and suppressed form. The activation or suppression function is caused by the binding of the DR3 response element as a heterodimer with the retinoid X receptor of the target genes. Due to the interactions with the basal transcriptional machinery and transcriptional cofactors, transcription is either activated or suppressed. When VDR is in its active form it regulates both phosphate and calcium metabolism, has immunosuppressive effects, and induces cell differentiation. When there are defects in the VDR that effect its metabolism it can lead to diseases such as severe rickets, secondary hyperparathyroidism, and hypocalcemia. Though defects in VDR can cause many diseases, fully functioning VDR can be used as treatment for disease such as cancer, autoimmune disease, psoriasis, osteoporosis, and renal osteodystrophy.[1] | ||
| Line 18: | Line 18: | ||
==Overall Structure== | ==Overall Structure== | ||
| - | The | + | The vitamin d receptor contains 427 amino acids with a total molecular weight of 48,289 Da. The protein is also composed almost entirely of alpha helices with only a single beta sheet. The vitamin D receptor also does not have a quaternary structure [2]. Vitamin D3 is a large organic compound made up of 27 carbon atoms, 44 hydrogen atoms and a single oxygen atom, with the ligand having a total molecular weight of 385 Da [3]. In studying the vitamin d receptor, the regions of the protein have been categorized into domains, with the A/B domain located at the N-terminus, the C domain, which is located between amino acid 20 and amino acid 115, the D domain, which is located between the end of the C domain and amino acid 220, and the EF domain, which encompasses the rest of the protein [4]. |
| + | <br> | ||
Other aspects of interest about the vitamin D receptor include the protein revealing a binding pocket when it is in its active folded state, allowing the ligand to bind to the receptor. The ligand interacts with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. The activation ligand is a nuclear receptor. There is also some empty space observed around the aliphatic chain [1]. | Other aspects of interest about the vitamin D receptor include the protein revealing a binding pocket when it is in its active folded state, allowing the ligand to bind to the receptor. The ligand interacts with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. The activation ligand is a nuclear receptor. There is also some empty space observed around the aliphatic chain [1]. | ||
The protein has an active conformation of 1,25 (OH)_2D_3 that has a ligand binding pocket in its active folded state. The activation ligand, a nuclear receptor (VDR), interacts with with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. There is some empty space observed around the aliphatic chain, indicating the presence of water to stabilize all possible hydrogen bonds [1]. | The protein has an active conformation of 1,25 (OH)_2D_3 that has a ligand binding pocket in its active folded state. The activation ligand, a nuclear receptor (VDR), interacts with with the activation helix by stabilizing the agonist position. This is accomplished through Van der Waals interactions between the ligand and the activation helix. There is some empty space observed around the aliphatic chain, indicating the presence of water to stabilize all possible hydrogen bonds [1]. | ||
| Line 62: | Line 63: | ||
3. Vitamin D3 https://pubchem.ncbi.nlm.nih.gov/compound/Vitamin_D3#section=2D-Structure (accessed Apr 10, 2016). | 3. Vitamin D3 https://pubchem.ncbi.nlm.nih.gov/compound/Vitamin_D3#section=2D-Structure (accessed Apr 10, 2016). | ||
| - | 4. Vitamin D Receptor | + | 4. Strugnell, S.; Deluca, H. The Vitamin D Receptor - Structure and Transcriptional Activation. Experimental Biology and Medicine1997, 215, 223–228. |
| - | 5. 3D Binding Pocket http://www.rcsb.org/pdb/explore/jmol.do?structureId=1DB1&residueNr=VDX (accessed Apr 4, 2016). | + | 5. Vitamin D Receptor http://pdb101.rcsb.org/motm/155 (accessed Apr 4, 2016). |
| + | |||
| + | 6. 3D Binding Pocket http://www.rcsb.org/pdb/explore/jmol.do?structureId=1DB1&residueNr=VDX (accessed Apr 4, 2016). | ||
Revision as of 21:42, 10 April 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Vitamin D receptor/vitamin D (1db1)[1]
by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole
Student Projects for UMass Chemistry 423 Spring 2016
| |||||||||||