Introduction
Potatinib was developed as a treatment option for chronic myeloid leukemia (CML) as other inhibitors in treatment have become ineffective. BCR-ABL is a kinase with cancerous genetic mutation in chromosome 22 that leaves it always active. Further mutations in BCR-ABL has left earlier drugs of tyrosine kinases unable to bind in almost 30% of cases over a course of five years of treatment. The newer, mutant BCR-ABL kinase’s ability to develop new resistances has pushed for newer developments in inhibitors, such as Potatinib[2].
Fibroblast growth factor (FGFR) signaling is the factor that normally activates the BCR-ABL kinase. Also, it is the protein behind both tissue development and repair, the disruption of which leads to tumor growth. The activation of BCR-ABL happens through a series of cascading signals that induce proliferation and migration in cells. Though mutations in the regulation of the FGFR tyrosine kinase family can result in malignant tumor growth[3]. The tyrosine kinase inhibitor Ponatinib has been used to to the mutant version of kinase BCR-ABL by the enzyme's specific "DFG-out" conformation (in turquoise). This conformation has the phenylalanine group of BCR-ABL flipped out of its hydrophobic binding site. Ponatinib is the first of its kind to be able to inhibit this specific mutation in BCR-ABL of the "DGF-out" combination[3].
The side effects have caused Ponatinib to fall under scrutiny from the U.S. Food and Drug Administration (FDA). It has shown to increase chances of deadly blood clotting and restenosis in both arteries and veins with a rate of about 1 in 5 patients. The drug has also shown to increase risk of heart attack and overall worsening of heart disease in patients[2].
Overall Structure
In terms of , FGFR in complex with Ponatinib consists of two domains, which is the characteristic structure exhibited by kinases. The N-terminal domain is the smaller of the two, and it contains a five-stranded beta sheet and an alpha carbon helix. The larger C-terminal domain is primarily alpha helical in structure. The alpha helices are shown in fuchsia and the beta strands are shown in orange. A hinge links the two regions. A network of hydrogen-bonds between three conserved residues – Glu551, Asn535, and Lys627 – exists in the hinge region. This hydrogen-bonding controls the kinase activity of FGFR.
In its active form, FGFR is dimerized and contains two activated intracellular substrates. The binding of a coreceptor, β-Klotho, stabilizes the activated complex. A DFG moiety is found in BCR-ABL, the conformation of which plays a key role in binding Ponatinib. Another defining feature of active FGFR is its . Four residues in the spine – Leu536, Met524, His610, and Phe631 (in orange) – are highly conserved. A gatekeeper residue is present at the beginning of the hinge, and interactions among the four hydrophobic spine residues link the gatekeeper to Tyr643 in the activation loop. This activation loop is glycine-rich and found in the kinase domain of FGFR[1].
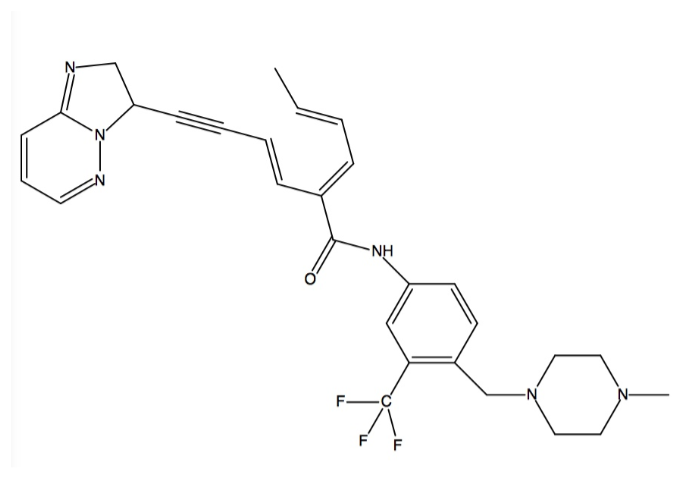
The structure of Ponatinib is shown as follows:
Binding Interactions
Kinases are the largest drug targets currently being tested in clinical trials. All kinases possess a biolobal fold that is a smaller N-terminal and a larger C-terminal lobe joined together by a “hinge.” The cofactor ATP binds deeply into a pocket between the lobes and binds to the hinge region. The imposition of any other residue in this ATP-binding pocket controls access to the hydrophobic pocket by separating the adenine binding site from an adjacent hydrophobic pocket. Such residues are termed “gatekeepers,” and are critical considerations in the development of drugs to treat CML because of the mutations that these residues can ensue. Gatekeeper mutations that convert a small hydrophilic residue into a large hydrophobic residue are one example of what has been shown to result in drug resistance, specifically to the most well-known ABL inhibitors like imatinib (Gleevec)[4]. Ponatinib is a third generation type II pan-BCR-ABL kinase inhibitor, which allows it to bind even with the presence of gatekeeper mutations[5]. Type II inhibitors are classified by binding to the hydrophobic and allosteric pocket that is only accessible in the DFG-out conformation and that is next to the ATP binding pocket. Additionally, type II inhibitors extend deep into the adenine pocket and hydrogen bond with the hinge region[4]. This unique property is caused by ponatinib’s ability to overcome resistances of the BCR-ABL gatekeeper mutant T315I at low concentrations (low IC50s ranging from 0.5 nM to 36 nM) by an ethynyl linker in the conformation (see color chart below)[5] [6]. The T315I mutation accounts for 15-20% of all clinically observed mutations and it is resistant to all previous generation drugs (imatinib, nilotinib, dasatinib). Additionally, ponatinib has a very high potency against native ABL which allows the binding energy to be distributed over many protein residues[5].
.
The specific binding of ponatinib can be categorized into and explained by five major chemical components, they are: (1) the template that interacts with the hinge region; (2) the A ring that occupies the hydrophobic pocket behind the gatekeeper residue; (3) the key ethynyl linker that joins the template and A ring, and that interacts with the gatekeeper residue (linker 1); (4) the A-B ring linker (linker 2); and (5) the B ring that binds to the DFG-out pocket[5].
Ponatinib binds into the ATP binding pocket between the N and C lobes to induce a shift from the DFG-in to the DFG-out conformation. It covers an immense region that spans from the kinase (in cyan) region (back of kinase) to the catalytic pocket (front of kinase). Three sites are engaged in the ATP binding cleft by ponatinib’s aromatic rings. In the first site, the imidazo[1,2b]pyridazine scaffold takes up the same space as the adenine ring of ATP and it is able to form one with the backbone amide nitrogen atom of Ala553 (in magenta) in the hinge [2]. Both of its rings form several Van der Waals contacts with residues in the N and C lobes of the adenine binding site as well[5]. Rigid acetylene linkage drives the rest of the drug into the back of the ATP binding pocket. In the second site, the methylphenyl group displaces the side chain of the catalytic Lys503 and its aromatic ring binds to the hydrophobic pocket that is formed by Val550, the residue (in yellow), and Met524[2]. Val550 is stabilized by the benzimide group[6]. This displacement allows Glu520 in the αC helix to hydrogen bond with the amide linkage between the aromatic rings in ponatinib. In the third site, Phe631 of DFG is expelled out of the cleft by ponatinib’s 3-trifluoromethylphenyl moiety, which takes the place of Phe631. Phe631’s new position enables it to make hydrophobic contact with the drug’s scaffold and acetylene linker. Also, Asp630 of DFG becomes available for hydrogen bonding with the amide linkage between the aromatic rings in ponatinib. This also puts the piperazine ring in the position to engage in hydrogen bonding with the catalytic loop. This is shift forms the DFG-out conformation[2].
Other inhibitors are not as potent as ponatinib against FGFR kinases because they are unable to penetrate far enough to access the third site and cannot assume the DFG-out conformation[2].
Additional Features
Ponatinib is an orally ingested tyrosine kinase inhibitor that has revealed successful avenues of treatment for counteracting the effects of angiogenesis in tumor growth. Besides the inhibition of FGFRs, this agent inhibits tyrosine kinases involved in vascular endothelial growth factor receptors. Ponatinib is considered a third generation TKI that can treat even the most drug-therapy resistant mutations that previous TKIs were incapable of treating[7].
The brand name for ponatinib is Iclusig. Iclusig received an accelerated approval grant through the Food and Drug Administration. It was mainly prescribed to patients suffering from Chronic Myeloid Leukemia or Acute Lymphoblastic Leukemia who did not make any progress with the first and second generation TKIs. However, the clinical trials data displayed a spike in adverse effects. These consequences include heart failure, stroke, coronary artery disease, loss of blood flow to body parts leading to amputation amongst other narrowing of blood vessels[8].
FGFR-4 is abundantly present in human prostate cancer observed in vitro and in mouse model simulations[9]. A of FGFR-4 with (Arg(388)) replacing (Gly(388)) is associated with increased human prostate cancer. This causes increased receptor stability and activation[10]. A study revealed that the inhibition of FGFR-4 signaling completely curtailed prostate cancer cell lines that were responsible for tumor growth[9]. Due to the significant results of diminished cell growth in treated tumors, targeting fibroblast growth factor signaling appears to provide a promising step towards combating aggressive prostate cancer.
Quiz Question 1
Ponatinib is unique in it's ability to bind to the mutated BCR-ABL because of it's preference to shift to the DFG-out conformation. In theory, if a competitive inhibitor was created by nature to prevent Ponatinib from binding to BCR-ABL to further its drug resistance, what specific structure characteristics would the inhibitor need to posses? Consider the unique binding methods of Ponatinib and the conformation.
a. Small, fully conjugated aromatic system with no electronegative substituents, to prevent unwanted hydrogen bonding.
b. Multiple ring system, one ring particularly for hydrogen bonding and another capable of binding in a hydrophobic pocket.
c. Polymer chain with an ester linkage and a hydroxyl end group .
d. Metal center that binds four large, nonpolar hydrocarbon ligands that exhibit significant steric hindrance.
See Also
Credits
Introduction - Emily & Cory*
Overall Structure - Nicole* & Connor
Drug Binding Site - Julie* & Cory
Additional Features - Connor* & Nicole
Quiz Question 1 - Julie & Emily*
References
- ↑ 1.0 1.1 Tucker JA, Klein T, Breed J, Breeze AL, Overman R, Phillips C, Norman RA. Structural Insights into FGFR Kinase Isoform Selectivity: Diverse Binding Modes of AZD4547 and Ponatinib in Complex with FGFR1 and FGFR4. Structure. 2014 Dec 2;22(12):1764-74. doi: 10.1016/j.str.2014.09.019. Epub 2014, Nov 20. PMID:25465127 doi:http://dx.doi.org/10.1016/j.str.2014.09.019
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Zhou T, Commodore L, Huang WS, Wang Y, Thomas M, Keats J, Xu Q, Rivera VM, Shakespeare WC, Clackson T, Dalgarno DC, Zhu X. Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance. Chem Biol Drug Des. 2011 Jan;77(1):1-11. doi:, 10.1111/j.1747-0285.2010.01054.x. Epub 2010 Nov 30. PMID:21118377 doi:10.1111/j.1747-0285.2010.01054.x
- ↑ Cite error: Invalid
<ref> tag;
no text was provided for refs named two
- ↑ 4.0 4.1 Huang Z, Tan L, Wang H, Liu Y, Blais S, Deng J, Neubert TA, Gray NS, Li X, Mohammadi M. DFG-out Mode of Inhibition by an Irreversible Type-1 Inhibitor Capable of Overcoming Gate-Keeper Mutations in FGF Receptors. ACS Chem Biol. 2014 Oct 27. PMID:25317566 doi:http://dx.doi.org/10.1021/cb500674s
- ↑ 5.0 5.1 5.2 5.3 5.4 Lesca E, Lammens A, Huber R, Augustin M. Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase. J Mol Biol. 2014 Sep 11. pii: S0022-2836(14)00474-4. doi:, 10.1016/j.jmb.2014.09.004. PMID:25219510 doi:http://dx.doi.org/10.1016/j.jmb.2014.09.004
- ↑ 6.0 6.1 Vijayan RS, He P, Modi V, Duong-Ly KC, Ma H, Peterson JR, Dunbrack RL Jr, Levy RM. Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated type II inhibitors. J Med Chem. 2015 Jan 8;58(1):466-79. doi: 10.1021/jm501603h. Epub 2014 Dec 12. PMID:25478866 doi:http://dx.doi.org/10.1021/jm501603h
- ↑ Price KE, Saleem N, Lee G, Steinberg M. Potential of ponatinib to treat chronic myeloid leukemia and acute lymphoblastic leukemia. Onco Targets Ther. 2013 Aug 20;6:1111-8. doi: 10.2147/OTT.S36980. eCollection, 2013. PMID:23986642 doi:http://dx.doi.org/10.2147/OTT.S36980
- ↑ FDA Drug Safety Communication: FDA investigating leukemia drug Iclusig (ponatinib) after increased reports of serious blood clots in arteries and veins; Drug Safety and Availability; United States Food and Drug Administration (2013). Web. [1]
- ↑ 9.0 9.1 Feng S, Shao L, Yu W, Gavine P, Ittmann M. Targeting fibroblast growth factor receptor signaling inhibits prostate cancer progression. Clin Cancer Res. 2012 Jul 15;18(14):3880-8. doi: 10.1158/1078-0432.CCR-11-3214., Epub 2012 May 9. PMID:22573348 doi:http://dx.doi.org/10.1158/1078-0432.CCR-11-3214
- ↑ Wang J, Yu W, Cai Y, Ren C, Ittmann MM. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008 Aug;10(8):847-56. PMID:18670643

