We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 425
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
[[Student Projects for UMass Chemistry 423 Spring 2016]] | [[Student Projects for UMass Chemistry 423 Spring 2016]] | ||
| - | <StructureSection load='4uxq' size='350' side='right' caption=' | + | <StructureSection load='4uxq' size='350' side='right' caption='FGFR in complex with Ponatinib is a highly effective inhibitory treatment for CML ([[4uxq]])' scene=''> |
==Introduction== | ==Introduction== | ||
| Line 15: | Line 15: | ||
Fibroblast growth factor (FGFR) signaling is the factor that normally activates the BCR-ABL kinase. Also, it is the protein behind both tissue development and repair, the disruption of FGFR leads to tumor growth. The activation of BCR-ABL happens through a series of cascading signals that induce proliferation and migration in cells. Mutations in the regulation of the FGFR tyrosine kinases can be diresctly correlated to malignant tumor growth<ref name="one" />. The tyrosine kinase inhibitor Ponatinib has been used to <scene name='48/483882/Activation_loop/1'>bind</scene> to the mutant version of kinase BCR-ABL by the enzyme's specific "DFG-out" conformation (in <font color='turquoise'><b>turquoise</b></font>). This conformation has the phenylalanine group of BCR-ABL flipped out of its hydrophobic binding site. Ponatinib is the first of its kind to be able to inhibit this specific mutation in BCR-ABL of the "DGF-out" conformation<ref name="seven">PMID: 21118377 </ref>. | Fibroblast growth factor (FGFR) signaling is the factor that normally activates the BCR-ABL kinase. Also, it is the protein behind both tissue development and repair, the disruption of FGFR leads to tumor growth. The activation of BCR-ABL happens through a series of cascading signals that induce proliferation and migration in cells. Mutations in the regulation of the FGFR tyrosine kinases can be diresctly correlated to malignant tumor growth<ref name="one" />. The tyrosine kinase inhibitor Ponatinib has been used to <scene name='48/483882/Activation_loop/1'>bind</scene> to the mutant version of kinase BCR-ABL by the enzyme's specific "DFG-out" conformation (in <font color='turquoise'><b>turquoise</b></font>). This conformation has the phenylalanine group of BCR-ABL flipped out of its hydrophobic binding site. Ponatinib is the first of its kind to be able to inhibit this specific mutation in BCR-ABL of the "DGF-out" conformation<ref name="seven">PMID: 21118377 </ref>. | ||
| - | + | Ponatinib's harmful side effects have caused it to fall under scrutiny from the U.S. Food and Drug Administration (FDA). It has shown to increase chances of deadly blood clotting and restenosis in both arteries and veins with a rate of about 1 in 5 patients. The drug has also shown to increase risk of heart attack and overall worsening of heart disease in patients<ref name="seven" />. | |
| Line 28: | Line 28: | ||
==Binding Interactions== | ==Binding Interactions== | ||
| - | Kinases are the largest drug targets being tested. All kinases possess a biolobal fold that | + | |
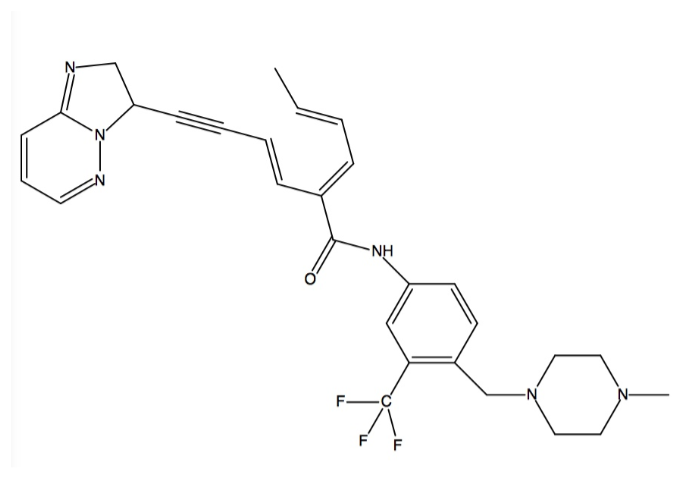
| + | Kinases are the largest drug targets currently being tested in clinical trials. All kinases possess a biolobal fold that is a smaller N-terminal and a larger C-terminal lobe joined together by a “hinge.” The cofactor ATP binds deeply into a pocket between the lobes and binds to the hinge region. The imposition of any other residue in this ATP-binding pocket controls access to the hydrophobic pocket by separating the adenine binding site from an adjacent hydrophobic pocket. Such residues are termed “gatekeepers,” and are critical considerations in the development of drugs to treat CML because of the mutations that these residues can ensue. Gatekeeper mutations that convert a small hydrophilic residue into a large hydrophobic residue are one example of what has been shown to result in drug resistance, specifically to the most well-known ABL inhibitors like imatinib (Gleevec)<ref name="four">PMID: 25317566</ref>. Ponatinib is a third generation type II pan-BCR-ABL kinase inhibitor, which allows it to bind even with the presence of gatekeeper mutations<ref name="five">PMID: 25219510</ref>. Type II inhibitors are classified by binding to the hydrophobic and allosteric pocket that is only accessible in the DFG-out conformation and that is next to the ATP binding pocket. Additionally, type II inhibitors extend deep into the adenine pocket and hydrogen bond with the hinge region<ref name="four" />. This unique property is caused by ponatinib’s ability to overcome resistances of the BCR-ABL gatekeeper mutant T315I at low concentrations (low IC50s ranging from 0.5 nM to 36 nM) by an ethynyl linker in the <scene name='48/483882/Active_sitezoom/1'>DFG-out</scene> conformation (see color chart below)<ref name="five" /> <ref name="six" />. The T315I mutation accounts for 15-20% of all clinically observed mutations and it is resistant to all previous generation drugs (imatinib, nilotinib, dasatinib). Additionally, ponatinib has a very high potency against native ABL which allows the binding energy to be distributed over many protein residues<ref name="five" />. | ||
<center><big>{{Template:ColorKey_N52C3Rainbow}}.</big></center> | <center><big>{{Template:ColorKey_N52C3Rainbow}}.</big></center> | ||
| - | + | The specific binding of ponatinib can be categorized into and explained by five major chemical components, they are: (1) the template that interacts with the hinge region; (2) the A ring that occupies the hydrophobic pocket behind the gatekeeper residue; (3) the key ethynyl linker that joins the template and A ring, and that interacts with the gatekeeper residue (linker 1); (4) the A-B ring linker (linker 2); and (5) the B ring that binds to the DFG-out pocket<ref name="five" />. | |
| + | |||
| + | Ponatinib binds into the ATP binding pocket between the N and C lobes to induce a shift from the DFG-in to the DFG-out conformation. It covers an immense region that spans from the kinase <scene name='48/483882/Hinge/1'>hinge</scene> (in <font color='cyan'><b>cyan</b></font>) region (back of kinase) to the catalytic pocket (front of kinase). Three sites are engaged in the ATP binding cleft by ponatinib’s aromatic rings. In the first site, the imidazo[1,2b]pyridazine scaffold takes up the same space as the adenine ring of ATP and it is able to form one <scene name='48/483882/Hingehbond/1'>hydrogen bond</scene> with the backbone amide nitrogen atom of Ala553 (in <font color='magenta'><b>magenta</b></font>) in the hinge <ref name="seven" />. Both of its rings form several Van der Waals contacts with residues in the N and C lobes of the adenine binding site as well<ref name="five" />. Rigid acetylene linkage drives the rest of the drug into the back of the ATP binding pocket. In the second site, the methylphenyl group displaces the side chain of the catalytic Lys503 and its aromatic ring binds to the hydrophobic pocket that is formed by Val550, the <scene name='48/483882/Gatekeeper/1'>gatekeeper</scene> residue (in <font color='yellow'><b>yellow</b></font>), and Met524<ref name="seven" />. Val550 is stabilized by the benzimide group<ref name="six">PMID: 25478866</ref>. This displacement allows Glu520 in the αC helix to hydrogen bond with the amide linkage between the aromatic rings in ponatinib. In the third site, Phe631 of DFG is expelled out of the cleft by ponatinib’s 3-trifluoromethylphenyl moiety, which takes the place of Phe631. Phe631’s new position enables it to make hydrophobic contact with the drug’s scaffold and acetylene linker. Also, Asp630 of DFG becomes available for hydrogen bonding with the amide linkage between the aromatic rings in ponatinib. This also puts the piperazine ring in the position to engage in hydrogen bonding with the catalytic loop. This is shift forms the DFG-out conformation<ref name="seven" />. | ||
| + | |||
| + | Other inhibitors are not as potent as ponatinib against FGFR kinases because they are unable to penetrate far enough to access the third site and cannot assume the DFG-out conformation<ref name="seven" />. | ||
==Additional Features== | ==Additional Features== | ||
Revision as of 01:37, 11 April 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Fibroblast Growth Factor Receptor/Ponatinib (4uxq) [1]
by Julie Boshar, Emily Boyle, Nicole Kirby, Cory Thomas, Connor Walsh
Student Projects for UMass Chemistry 423 Spring 2016
| |||||||||||