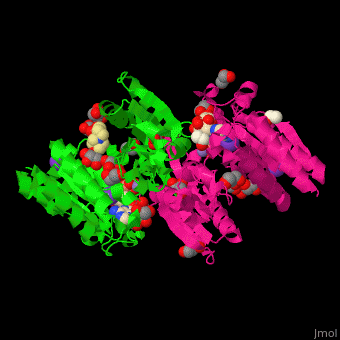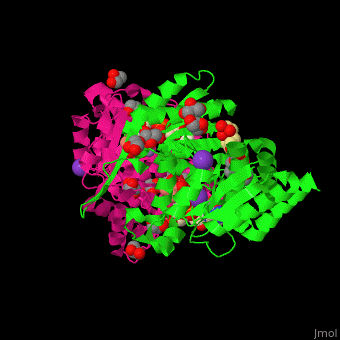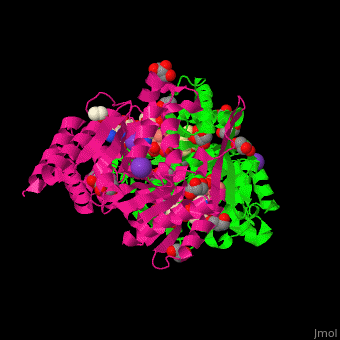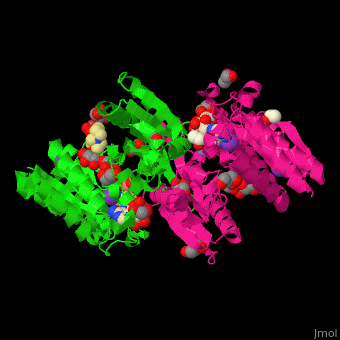Isopropylmalate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
| + | <scene name='49/490899/Cv/2'>3-isopropylmalate dehydrogenase complex with 3-isopropylmalate, glycerol, NAD, PMSF, Mn+2 and K- ions</scene>. | ||
| + | |||
IMDH active site containing the NAD cofactor is located between 2 subunits and contains the substrate and Mn+2 and K- ions<ref>PMID:25211160</ref>. | IMDH active site containing the NAD cofactor is located between 2 subunits and contains the substrate and Mn+2 and K- ions<ref>PMID:25211160</ref>. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:08, 11 April 2016
| |||||||||||
3D structures of isopropylmalate dehydrogenase
Updated on 11-April-2016
References
- ↑ Martignon S, Rossi F, Rizzi M. Expression, purification and characterisation of Haemophilus influenzae 3-isopropylmalate dehydrogenase (LeuB). Protein Pept Lett. 2007;14(8):822-7. PMID:17979826
- ↑ Pallo A, Olah J, Graczer E, Merli A, Zavodszky P, Weiss MS, Vas M. Structural and energetic basis of isopropylmalate dehydrogenase enzyme catalysis. FEBS J. 2014 Sep 11. doi: 10.1111/febs.13044. PMID:25211160 doi:http://dx.doi.org/10.1111/febs.13044