Hyaluronidase
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
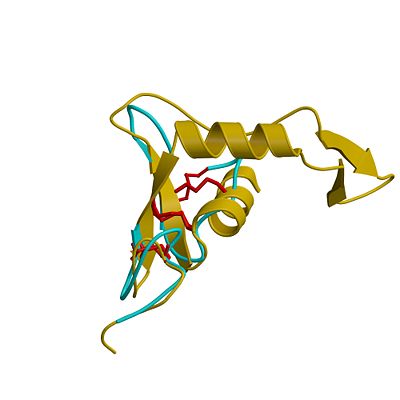
[[Image:hya_xdt_100906.jpeg|left|thumb|'''Superposition of the EGF-like domain of hyaluronidase-1 (yellow) and the heparin-binding EGF-like growth factor (light blue)'''. The three disulphide bonds of the EGF domains (highlighted red) exhibit the same pattern in the primary structure and are located in similar positions in the 3D structure. |400px]] | [[Image:hya_xdt_100906.jpeg|left|thumb|'''Superposition of the EGF-like domain of hyaluronidase-1 (yellow) and the heparin-binding EGF-like growth factor (light blue)'''. The three disulphide bonds of the EGF domains (highlighted red) exhibit the same pattern in the primary structure and are located in similar positions in the 3D structure. |400px]] | ||
{{Clear}} | {{Clear}} | ||
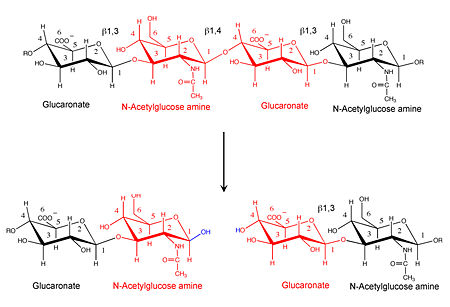
| - | [[Image:Hyal reaction.jpg|thumb|left|'''Hyaluronidase cleaves the β1,4-glycosidic bond of the glycosaminoglycan hyaluronan''' | | + | [[Image:Hyal reaction.jpg|thumb|left|'''Hyaluronidase cleaves the β1,4-glycosidic bond of the glycosaminoglycan hyaluronan''' |450px]] |
{{Clear}} | {{Clear}} | ||
== Relevance == | == Relevance == | ||
| Line 13: | Line 13: | ||
== Structural highlights == | == Structural highlights == | ||
HU structure contains an <scene name='49/497055/Cv/2'>N-terminal, linker and C-terminal domains</scene>. The active site is in a cleft in the N-terminal domain. Two HUA molecules are bound in the active site making contacts with some HU residues and multiple contacts with water molecules<ref>PMID:10843845</ref>. | HU structure contains an <scene name='49/497055/Cv/2'>N-terminal, linker and C-terminal domains</scene>. The active site is in a cleft in the N-terminal domain. Two HUA molecules are bound in the active site making contacts with some HU residues and multiple contacts with water molecules<ref>PMID:10843845</ref>. | ||
| + | |||
| + | <scene name='49/497055/Cv/5'>1st active site of HU</scene>. Water molecules shown as red spheres. | ||
| + | |||
| + | <scene name='49/497055/Cv/6'>2nd active site of HU</scene>. | ||
| + | |||
| + | <scene name='49/497055/Cv/7'>Dimethylarsinate binding site</scene>. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 08:34, 12 April 2016
| |||||||||||
3D structures of hyaluronidase
Updated on 12-April-2016
Reference
- ↑ Chao KL, Muthukumar L, Herzberg O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry. 2007 Jun 12;46(23):6911-20. Epub 2007 May 16. PMID:17503783 doi:10.1021/bi700382g
- ↑ Ponnuraj K, Jedrzejas MJ. Mechanism of hyaluronan binding and degradation: structure of Streptococcus pneumoniae hyaluronate lyase in complex with hyaluronic acid disaccharide at 1.7 A resolution. J Mol Biol. 2000 Jun 16;299(4):885-95. PMID:10843845 doi:10.1006/jmbi.2000.3817
Created with the participation of Osnat Herzberg, Eran Hodis, Joel L. Sussman, Jaime Prilusky.