We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Nos1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
This is a default text for your page '''Nos1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Nos1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| + | |||
| + | == Location == | ||
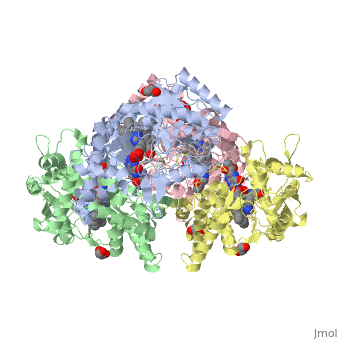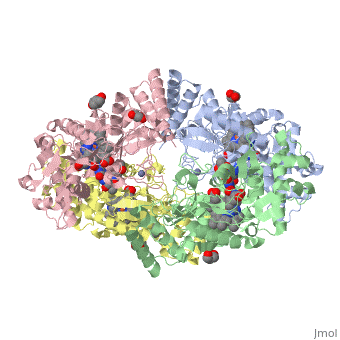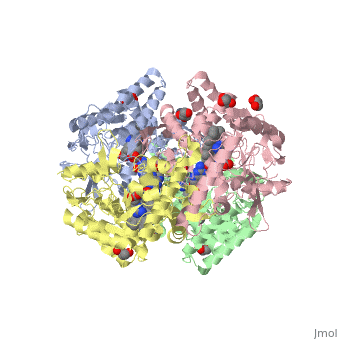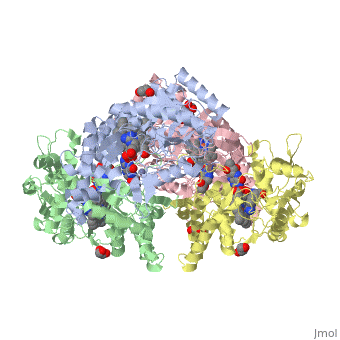
| + | The human gene NOS1, found on chromosome 12, encodes for a nitric oxide synthase. Nitric oxide synthases consume L-arginine to produce nitric oxide, which is used as a signaling molecule. The chemical equation is shown below. | ||
| + | |||
| + | 2 L-arginine + 3 NADPH + 4 O(2) = 2 L- citrulline + 2 nitric oxide + 3 NADP(+) + 4 H(2)O | ||
| + | |||
| + | This gene is expressed by all cell types because nitric oxide is used in a wide variety of processes including heme binding, NADP binding, calcium signaling, and oxidation-reduction reactions. | ||
== Function == | == Function == | ||
== Disease == | == Disease == | ||
| - | Nitric oxide (NO) is produced from one of three synthases present within the body: neuronal nitric oxide synthase 1 (NOS1), inducible nitric oxide synthase 2 (NOS2), and endothelial nitric oxide synthase 3 (NOS3) (Shinkai, et. al, 2002). Specifically, NOS1 has nine different first exons that lead to multiple NOS1 transcripts with different 5’-untranslated regions (Galimberti, et. al, 2008). An advantage of having multiple first exons that can be alternatively spliced and expressed is that NOS1 can be specific and specifically regulated for different tissues (Galimberti, et. al, 2008). A disadvantage to having multiple first exons is that there is a higher possibility of mutations, which would affect NO production and thus have an effect on the second messenger cyclic guanine monophosphate (cGMP) production (Shinkai, et. al, 2002). Furthermore, because NO is an oxyradical, overproduction caused by a mutation can lead to neural tissue damage (Shinkai, et. al, 2002). Numerous pathologies may arise from neural tissue damage, and one study suggested overproduction of NO that leads to such damage is possibly an influential factor in developing schizophrenia (Shinkai, et. al, 2002). | ||
| - | Single nucleotide polymorphisms (SNPs) and various lengths of tandem repeats have been linked in other disorders of the brain such as Alzheimer’s and Parkinson’s diseases (Galimberti, et. al, 2008; Rife, et. al, 2009). Out of three identified SNPs occurring in alternative exon 1c, only the SNP G-84A has a functional effect that decreases transcription levels (Galimberti, et. al, 2008). However, various lengths of tandem repeats present in the alternative exon 1f has been shown to be a potential factor in both Alzheimer’s and Parkinson’s diseases (Galimberti, et. al, 2008; Rife, et. al, 2009). Shorter tandem TG repeats are possibly associated with the development of Alzheimer’s disease, and longer tandem TG repeats are possibly associated with the development of Parkinson’s disease (Rife, et. al, 2009). Although schizophrenia, Alzheimer’s, and Parkinson’s diseases have genetic influences, mutations in NOS1 can be a risk indicator for developing these diseases (Shinkai, et. al, 2002; Galimberti, et. al, 2008; Rife, et. al, 2009). | ||
== Relevance == | == Relevance == | ||
| Line 19: | Line 24: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | Galimberti, D., Scarpini, E., Venturelli, E., Strobel, A., Herterich, S., Fenogolio, C., Guidi, I., Scalabrini, D., Cortini, F., Bresolin, N., Lesch, K., and Reif, A. (2008) Association of a NOS1 promoter repeat with Alzheimer’s disease. Neurobiology of Aging. 29, 1359-1365. doi:10.1016/j.neurobiolaging.2007.03.003 | ||
| - | Rife, T., Rasoul, B., Pullen, N., Mitchell, D., Grathwol, K., and Kurth, J. (2009) The effect of a promoter polymorphism on transcription of nitric oxide synthase 1 and its relevance to Parkinson’s disease. Journal of Neuroscience Research. 87, 2319-2325. doi:10.1005/jnr.22045 | ||
| - | Shinkai, T., Ohmori, O., Hori, H., and Nakamura, J. (2002) Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Molecular Psychiatry. 7, 560-563. doi:10.1038/sj.mp.4001041 | ||
<references/> | <references/> | ||
Revision as of 17:45, 12 April 2016
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644