This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
TEM1 Class Antibiotic Resistance Proteins
From Proteopedia
| Line 25: | Line 25: | ||
== Function == | == Function == | ||
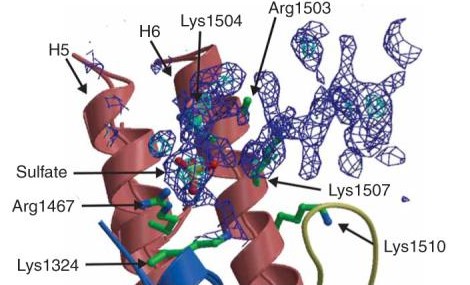
| - | + | There are two domains of this protein with the catalytic cleft in the shared region between the two domains. Mutations that occur in the catalytic region are able to slow down the catalysis that inactivates the antibiotics. | |
== Disease == | == Disease == | ||
| - | == Relevance | + | == Relevance = |
| - | + | The frequency of mutations in bacteria make it an evolving resistance. The ability of TEM1 CLass Antibiotic Resistant protein to selectively mutate a single amino acid to inhibit the beta lactamase allows for a more adaptable resistance source. | |
== Structural highlights == | == Structural highlights == | ||
Revision as of 10:59, 14 April 2016
| |||||||||||
References
1. Davies, J.; Davies, G. Origins and Evolution of Antibiotic Resistance. Microbiol Mol Biol Rev. 2010, Sep; 74(3): 417–433.
2. National Institute of Health. Stop the Spread of Superbugs Help Fight Drug-Resistant Bacteria. https://newsinhealth.nih.gov/issue/feb2014/feature1. (Last accessed: April 11, 2016).
3. Dablon et al. The catalytic mechanism of f3-lactamases: NMR titration of an active-site lysine residue of the TEM-1 enzyme. Proc. Natl. Acad. Sci. USA. 1996, 74: 1747-1752.
4. Fonze, E.; Charlier, P.; To'th, Y.; Vermeire, M.; Raquet, X.; Dubus, A.; Frere, J. M. TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant.
Acta Crystallogr. D Biol. Crystallogr. 1995, 51, 682-694.
5. Lenfant, F.; Labia, R.; Masson, J. -. Replacement of lysine 234 affects transition state stabilization in the active site of ß-lactamase TEM1. J. Biol. Chem. 1991, 266, 17187-17194.
Proteopedia Page Contributors and Editors (what is this?)
Kenna Salvatore, Matt O'Malley, Ryan Hunter Wilson, Riley Culhane, Michal Harel