We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1162
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='4oo9' size='340' side='right' caption='metabotropic Glutamate Receptor 5 PDB:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]' scene='72/726409/Overview/ | + | <StructureSection load='4oo9' size='340' side='right' caption='metabotropic Glutamate Receptor 5 PDB:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]' scene='72/726409/Overview/6'> |
= metabotropic Glutamate Receptor 5 = | = metabotropic Glutamate Receptor 5 = | ||
== Introduction == | == Introduction == | ||
| - | G-coupled protein receptors [https://en.wikipedia.org/wiki/G_protein–coupled_receptor (GPCR's)] are helical trans-membrane proteins that bind to an extracellular signal and activate a cellular response. The human genome encodes for approximately 750 GPCR's, 350 of which are known to respond to extracellular ligands.<ref name="GPCRRep">PMID: 12679517 </ref> GPCR's are divided into four major classes based on sequence similarity and transduction mechanism: Class A,B,C, and F.<ref name="MSGPCR">PMID:23407534</ref> Metabotropic Glutamate Receptor 5 (<scene name='72/726409/Overview/ | + | G-coupled protein receptors [https://en.wikipedia.org/wiki/G_protein–coupled_receptor (GPCR's)] are helical trans-membrane proteins that bind to an extracellular signal and activate a cellular response. The human genome encodes for approximately 750 GPCR's, 350 of which are known to respond to extracellular ligands.<ref name="GPCRRep">PMID: 12679517 </ref> GPCR's are divided into four major classes based on sequence similarity and transduction mechanism: Class A,B,C, and F.<ref name="MSGPCR">PMID:23407534</ref> Metabotropic Glutamate Receptor 5 (<scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene>) is a class C GPCR that is involved in the G<sub>q</sub> pathway.<ref name="CCGPCR">PMID:12782243</ref> In this pathway, glutamate binds to the extracellular domain of mGlu<sub>5</sub>, and the trans-membrane domain then undergoes a conformational change that activates the coupled [http://proteopedia.org/wiki/index.php/GTP-binding_protein G-protein] on the intracellular side of the membrane.<ref name="Primary">PMID: 25042998 </ref> The activated G-protein disassociates, and the alpha subunit activates [https://en.wikipedia.org/wiki/Phospholipase_C Phospholipase C]. Phospholipase C in turn cleaves [https://en.wikipedia.org/wiki/Phosphatidylinositol_4,5-bisphosphate PIP2] to [https://en.wikipedia.org/wiki/Diglyceride DAG] and [https://en.wikipedia.org/wiki/Inositol_trisphosphate IP3]. IP3 then binds to calcium channels on the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum Endoplasmic reticulum], creating an increased cellular concentration of calcium. Increased calcium concentrations thus lead to increased neuronal activity.<ref name="MSGPCR">PMID:23407534</ref> Due to its involvement in neuronal activity, mGlu<sub>5</sub> is highly expressed in neuronal and glial cells in the central nervous system, where glutamate serves as the major neurotransmitter. |
== Structure == | == Structure == | ||
=== Overall Stucture === | === Overall Stucture === | ||
| - | <scene name='72/726409/Overview/ | + | <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> is seen as a [https://en.wikipedia.org/wiki/Protein_dimer homodimer] ''in vivo,'' with each subunit being comprised of three domains: extracellular, trans-membrane and cysteine-rich. mGlu<sub>5</sub> is centered on the trans-membrane domain, comprised of seven α-helices all roughly parallel.<ref name="Primary">PMID: 25042998 </ref> Also displayed is Intracellular Loop (ICL) 1 which forms a short α-helix and interacts directly with a trans-membrane helix to stabilize mGlu<sub>5</sub>’s conformation. Additionally, ICL3 and Extracellular Loops (ECL) 1 and 3 all lack secondary structure. and ECL2 interacts with trans-membrane (TM) helices 1, 2, and 3 as well as ECL 1, again helping to stabilize the protein’s overall conformation.<ref name="Primary">PMID: 25042998 </ref> |
===Key Interactions=== | ===Key Interactions=== | ||
| - | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/ | + | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> being represented in the inactivate state. While in the inactive state, glutamate binding to mGlu<sub>5</sub> triggers a conformational change that leads to mGlu<sub>5</sub> being in the active state and hence initiates the aforementioned [https://en.wikipedia.org/wiki/Gq_alpha_subunit G<sub>q</sub> pathway]. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/2'>Ionic Lock</scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately substituted with alanine, resulting in constitutive activity of the GPCR and its coupled pathway.<ref name="Primary">PMID: 25042998 </ref> A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/2'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 was substituted with alanine, high levels of activity were seen in the mutant GPCR.<ref name="Primary">PMID: 25042998 </ref> A <scene name='72/726404/Scene_6/10'>Disulfide Bond </scene> between Cysteine 644 of TM3 and Cysteine 733 of <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene> is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s.<ref name="Primary">PMID: 25042998 </ref> The ECL2's presence combined with the helical bundle of the trans-membrane domain creates a <scene name='72/726409/Electrogradient2/9'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as it binds to the extracellular domain, but this entrance dictates potential drug targets that act through allosteric modulation.<ref name="Primary">PMID: 25042998 </ref> |
== Clinical Relevance == | == Clinical Relevance == | ||
===Role in Diseases=== | ===Role in Diseases=== | ||
| Line 16: | Line 16: | ||
The structure of mGlu<sub>5</sub> bound to the NAM [https://en.wikipedia.org/wiki/Mavoglurant Mavoglurant] demonstrates how protein activity is decreased through drug interactions. | The structure of mGlu<sub>5</sub> bound to the NAM [https://en.wikipedia.org/wiki/Mavoglurant Mavoglurant] demonstrates how protein activity is decreased through drug interactions. | ||
<scene name='72/726404/Scene_7/6'>Mavoglurant</scene> binds within the allosteric binding site in the core of the seven trans-membrane α-helices, having passed through the restricted entrance formed by the <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene>. Bound Mavoglurant forms multiple interactions with the protein that further stabilize the inactive conformation. | <scene name='72/726404/Scene_7/6'>Mavoglurant</scene> binds within the allosteric binding site in the core of the seven trans-membrane α-helices, having passed through the restricted entrance formed by the <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene>. Bound Mavoglurant forms multiple interactions with the protein that further stabilize the inactive conformation. | ||
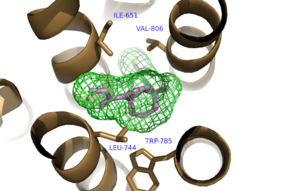
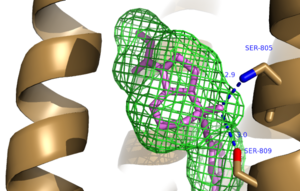
| - | The bicyclic ring system of the drug is surrounded by a pocket of mainly hydrophobic residues including Val 806, Met 802, Phe 788, Trp 785, Leu 744, Ile 651, Pro 655, and Asn 747 (Figure 1).<ref name="Primary">PMID: 25042998 </ref> The carbamate tail of Mavoglurant forms a hydrogen bond through its carbonyl oxygen to the amide side-chain of Asn 747 of TM4 | + | The bicyclic ring system of the drug is surrounded by a pocket of mainly hydrophobic residues including Val 806, Met 802, Phe 788, Trp 785, Leu 744, Ile 651, Pro 655, and Asn 747 (Figure 1).<ref name="Primary">PMID: 25042998 </ref> The carbamate tail of Mavoglurant forms a hydrogen bond through its carbonyl oxygen to the amide side-chain of Asn 747 of TM4. A hydroxyl group similarly forms hydrogen bonds to mGlu<sub>5</sub>, specifically at two serine residues (S805 and S809) of TM7 (Figure 2). These residues form a hydrogen bonding network to other residues through their main chain atoms and a coordinated water molecule. The interactions between Mavoglurant and mGlu<sub>5</sub> involve TM helices that were not previously stabilized by any strong interactions, introducing a new level of stability that favors the inactive conformation of the protein and hence decreases the overall activity of mGlu<sub>5</sub>.<ref name="Primary">PMID: 25042998 </ref> |
Mavoglurant was met by disappointing Fragile X-syndrome trial results and ultimately discontinued for Fragile X-syndrome in 2014, but testing for dyskinesia and [https://en.wikipedia.org/wiki/Obsessive%E2%80%93compulsive_disorder Obsessive-compulsive disorder] are still in progress. [https://en.wikipedia.org/wiki/Basimglurant Basimglurant] and [https://en.wikipedia.org/wiki/MTEP MTEP] are inhibitors that are similar to Mavoglurant as they both function as negative allosteric modulators to mGlu<sub>5</sub>, and they are currently under trials to serve as medications for depression.<ref name="Clinical">PMID: 26219727 </ref><ref name=”Clinical2”>PMID: 25043733 </ref> | Mavoglurant was met by disappointing Fragile X-syndrome trial results and ultimately discontinued for Fragile X-syndrome in 2014, but testing for dyskinesia and [https://en.wikipedia.org/wiki/Obsessive%E2%80%93compulsive_disorder Obsessive-compulsive disorder] are still in progress. [https://en.wikipedia.org/wiki/Basimglurant Basimglurant] and [https://en.wikipedia.org/wiki/MTEP MTEP] are inhibitors that are similar to Mavoglurant as they both function as negative allosteric modulators to mGlu<sub>5</sub>, and they are currently under trials to serve as medications for depression.<ref name="Clinical">PMID: 26219727 </ref><ref name=”Clinical2”>PMID: 25043733 </ref> | ||
Revision as of 17:39, 18 April 2016
| |||||||||||
References
- ↑ Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4903-8. Epub 2003 Apr 4. PMID:12679517 doi:http://dx.doi.org/10.1073/pnas.0230374100
- ↑ 2.0 2.1 Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013 Feb 14;494(7436):185-94. doi: 10.1038/nature11896. PMID:23407534 doi:http://dx.doi.org/10.1038/nature11896
- ↑ Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003 Jun;98(3):325-54. PMID:12782243
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 4.9 Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, Weir M, Wiggin GR, Marshall FH. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014 Jul 31;511(7511):557-62. doi: 10.1038/nature13396. Epub 2014 Jul 6. PMID:25042998 doi:http://dx.doi.org/10.1038/nature13396
- ↑ Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993 Nov 26;163(1):53-7. PMID:8295733
- ↑ 6.0 6.1 Li G, Jorgensen M, Campbell BM. Metabotropic glutamate receptor 5-negative allosteric modulators for the treatment of psychiatric and neurological disorders (2009-July 2013). Pharm Pat Anal. 2013 Nov;2(6):767-802. doi: 10.4155/ppa.13.58. PMID:24237242 doi:http://dx.doi.org/10.4155/ppa.13.58
- ↑ Fuxe K, Borroto-Escuela DO. Basimglurant for treatment of major depressive disorder: a novel negative allosteric modulator of metabotropic glutamate receptor 5. Expert Opin Investig Drugs. 2015;24(9):1247-60. doi:, 10.1517/13543784.2015.1074175. Epub 2015 Jul 29. PMID:26219727 doi:http://dx.doi.org/10.1517/13543784.2015.1074175
- ↑ Domin H, Szewczyk B, Wozniak M, Wawrzak-Wlecial A, Smialowska M. Antidepressant-like effect of the mGluR5 antagonist MTEP in an astroglial degeneration model of depression. Behav Brain Res. 2014 Oct 15;273:23-33. doi: 10.1016/j.bbr.2014.07.019. Epub 2014, Jul 18. PMID:25043733 doi:http://dx.doi.org/10.1016/j.bbr.2014.07.019