We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1165
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
Many of the [https://en.wikipedia.org/wiki/Residue_(chemistry) residues] in the binding pocket that are in direct contact with the glucagon molecule are [https://en.wikipedia.org/wiki/Ion charged] or are [https://en.wikipedia.org/wiki/Chemical_polarity polar]. | Many of the [https://en.wikipedia.org/wiki/Residue_(chemistry) residues] in the binding pocket that are in direct contact with the glucagon molecule are [https://en.wikipedia.org/wiki/Ion charged] or are [https://en.wikipedia.org/wiki/Chemical_polarity polar]. | ||
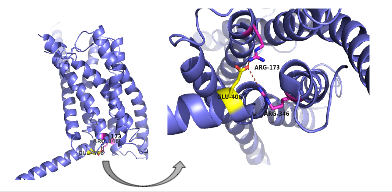
[[Image:Screen Shot 2016-03-29 at 3.24.43 PM.png|(|):|400 px|right|thumb|'''Figure 3: Salt Bridge'''. The salt bridge is located on the intracellular side at the bottom of the protein, in relation to the orientation it holds within the cell membrane. It is made between residues Glu 406, Arg 173, and Arg 346, as labeled in the figure.]] | [[Image:Screen Shot 2016-03-29 at 3.24.43 PM.png|(|):|400 px|right|thumb|'''Figure 3: Salt Bridge'''. The salt bridge is located on the intracellular side at the bottom of the protein, in relation to the orientation it holds within the cell membrane. It is made between residues Glu 406, Arg 173, and Arg 346, as labeled in the figure.]] | ||
| + | |||
There are also many smaller residues on glucagon that support the bulky residues on the GCGR. These residues are located within the binding pocket of the 7TM (Figure 4). <ref name="Ligands">PMID: 21542831</ref> There are specific amino acid interactions that hold the helices of the 7TM in the closed conformation that maximizes [http://www.chemicool.com/definition/affinity.html affinity]. <ref name="Ligands">PMID: 21542831</ref> This includes the [https://en.wikipedia.org/wiki/Disulfide disulfide bond] between Cys 294 and Cys 224 that was mentioned earlier that serves to hold the ECL1 and ECL2 in the proper orientation. Additionally, the [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] between Glu 406, Arg 173, and Arg 346, also mentioned earlier, hold the conformation together for higher affinity (Figure 3). <ref name="Ligands">PMID: 21542831</ref> Finally, alpha helical structure of the stalk is imperative to the affinity and binding of the glucagon. <ref name="Tips">PMID: 23863937</ref> | There are also many smaller residues on glucagon that support the bulky residues on the GCGR. These residues are located within the binding pocket of the 7TM (Figure 4). <ref name="Ligands">PMID: 21542831</ref> There are specific amino acid interactions that hold the helices of the 7TM in the closed conformation that maximizes [http://www.chemicool.com/definition/affinity.html affinity]. <ref name="Ligands">PMID: 21542831</ref> This includes the [https://en.wikipedia.org/wiki/Disulfide disulfide bond] between Cys 294 and Cys 224 that was mentioned earlier that serves to hold the ECL1 and ECL2 in the proper orientation. Additionally, the [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] between Glu 406, Arg 173, and Arg 346, also mentioned earlier, hold the conformation together for higher affinity (Figure 3). <ref name="Ligands">PMID: 21542831</ref> Finally, alpha helical structure of the stalk is imperative to the affinity and binding of the glucagon. <ref name="Tips">PMID: 23863937</ref> | ||
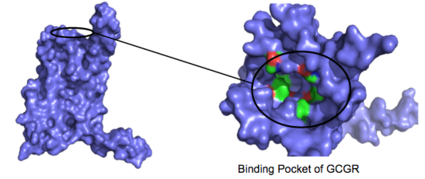
| - | [[Image:Screen_Shot_2016-03-22_at_5.28.03_PM.png|(|):|425 px|center|thumb|'''Figure 4: Binding Pocket Residues:''' side chains of carbon | + | [[Image:Screen_Shot_2016-03-22_at_5.28.03_PM.png|(|):|425 px|center|thumb|'''Figure 4: Binding Pocket Residues:''' Residues with side chains of carbon(utilizing the [https://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic effect] are shown in green and side chains containing oxygen ([https://en.wikipedia.org/wiki/Hydrophile hydrophilic] are shown in red. The properties of hydrophobicity and hydrophilicity of the residues create affinity for the binding of glucagon]] |
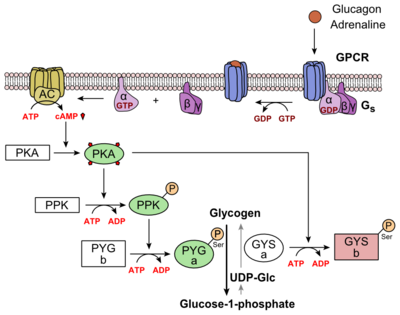
=Glucagon Signaling Pathway= | =Glucagon Signaling Pathway= | ||
Revision as of 22:12, 18 April 2016
References
- ↑ 1.0 1.1 Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ 3.0 3.1 3.2 3.3 Miller LJ, Dong M, Harikumar KG. Ligand binding and activation of the secretin receptor, a prototypic family B G protein-coupled receptor. Br J Pharmacol. 2012 May;166(1):18-26. doi: 10.1111/j.1476-5381.2011.01463.x. PMID:21542831 doi:http://dx.doi.org/10.1111/j.1476-5381.2011.01463.x
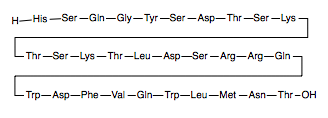
- ↑ Thomsen J, Kristiansen K, Brunfeldt K, Sundby F. The amino acid sequence of human glucagon. FEBS Lett. 1972 Apr 1;21(3):315-319. PMID:11946536
- ↑ 5.0 5.1 5.2 Bortolato A, Dore AS, Hollenstein K, Tehan BG, Mason JS, Marshall FH. Structure of Class B GPCRs: new horizons for drug discovery. Br J Pharmacol. 2014 Jul;171(13):3132-45. doi: 10.1111/bph.12689. PMID:24628305 doi:http://dx.doi.org/10.1111/bph.12689
- ↑ Mukund S, Shang Y, Clarke HJ, Madjidi A, Corn JE, Kates L, Kolumam G, Chiang V, Luis E, Murray J, Zhang Y, Hotzel I, Koth CM, Allan BB. Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor. J Biol Chem. 2013 Nov 4. PMID:24189067 doi:http://dx.doi.org/10.1074/jbc.M113.496984
- ↑ Hoare SR. Allosteric modulators of class B G-protein-coupled receptors. Curr Neuropharmacol. 2007 Sep;5(3):168-79. doi: 10.2174/157015907781695928. PMID:19305799 doi:http://dx.doi.org/10.2174/157015907781695928
- ↑ 8.0 8.1 8.2 Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859