We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1167
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
====Other Unique Structural Features ==== | ====Other Unique Structural Features ==== | ||
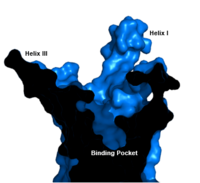
| - | An important interface stabilization interaction between Helices I and VII occurs between [https://en.wikipedia.org/wiki/Serine Ser] 152 of Helix I and Ser 390 of Helix VII. Due to their close proximity to one another, they form an important <scene name='72/721537/Ser-ser_hydrogen_bond/ | + | An important interface stabilization interaction between Helices I and VII occurs between [https://en.wikipedia.org/wiki/Serine Ser] 152 of Helix I and Ser 390 of Helix VII. Due to their close proximity to one another, they form an important <scene name='72/721537/Ser-ser_hydrogen_bond/3'>hydrogen bond</scene> which stabilizes the structure of GCGR. Mutations to the homologous residues Ser 135 and Ser 392 have been shown to alter receptor signaling in [https://en.wikipedia.org/wiki/Glucagon-like_peptide_1_receptor glucagon-like peptide-1 receptor] (GLP1R). |
Current revision
| This Sandbox is Reserved from Jan 11 through August 12, 2016 for use in the course CH462 Central Metabolism taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1160 through Sandbox Reserved 1184. |
To get started:
More help: Help:Editing |
Class B Human Glucagon G-Protein Coupled Receptor
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ 2.0 2.1 2.2 Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ 3.0 3.1 Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859
- ↑ 4.0 4.1 Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, Clarke HJ, Wong T, Chiang V, Luis E, Estevez A, Rondon J, Zhang Y, Hotzel I, Allan BB. Molecular basis for negative regulation of the glucagon receptor. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14393-8. Epub 2012 Aug 20. PMID:22908259 doi:http://dx.doi.org/10.1073/pnas.1206734109
- ↑ Mukund S, Shang Y, Clarke HJ, Madjidi A, Corn JE, Kates L, Kolumam G, Chiang V, Luis E, Murray J, Zhang Y, Hotzel I, Koth CM, Allan BB. Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor. J Biol Chem. 2013 Nov 4. PMID:24189067 doi:http://dx.doi.org/10.1074/jbc.M113.496984
- ↑ Mittermayer F, Caveney E, De Oliveira C, Gourgiotis L, Puri M, Tai LJ, Turner JR. Addressing unmet medical needs in type 2 diabetes: a narrative review of drugs under development. Curr Diabetes Rev. 2015;11(1):17-31. doi: 10.2174/1573399810666141224121927. PMID:25537454 doi:http://dx.doi.org/10.2174/1573399810666141224121927