Introduction
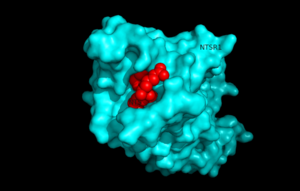
Figure 1.Top view of NTSR1 protein (blue) interacting with NTS ligand (red)
Neurotensin receptor 1 (NTSR1) is a G-protein coupled receptor (GPCR). GPCRs are a class of proteins with an extracellular binding domain and 7 transmembrane helices found only in eukaryotes that assist in propagating a cellular response. This is accomplished by the binding of molecules to the GPCR outside the cell,causing a conformational change and activating a signal transduction pathway via second messengers such as cyclic AMP, inositol triphosphate, and diacylglycerol.[1]. NTSR1 binds to the 13 amino acid peptide, neurotensin (NTS)[2], and the majority of the effects of NTS are mediated through NTSR1[2]. NTS has a variety of biological activities including a role in the leptin signaling pathways [3], tumor growth [4], and dopamine regulation [5].Recently NTSR1 was crystallized bound with the C-terminus of its tridecapeptide ligand, . The shortened ligand was used because it has a higher potency and efficacy than its full-length counterpart[2]. Class A GPCRs bind their ligands within the transmembrane core in a ligand binding pocket. The in NTSR1 is located at the top of the protein (Figure 1). NTSR1 also contains an allosteric . The Na+ binding pocket is located directly beneath the ligand binding pocket and the two pockets are separated by the residue Trp321. [1]. NTSR1 has been mutated to exist in both and states. This has led to a greater understanding of the structure of NTSR1 and how the structure influences its function.
Structure
Ligand Binding Pocket
On the extracellular side of the protein is the
. [2]
One key residue in this pocket is a Phenylalanine at position 358, which takes part in a network of hydrophobic stacking interactions. Cite error: Invalid <ref> tag;
refs with no content must have a name These interactions stabilize the Trp321 and Tyr324 residues allowing Tyr324 to interact with the C-terminal
via Van der Waals interactions .[2][1]
Without the hydrophobic stacking interactions that are facilitated by the Phe358, this binding interaction would be destabilized. Trp321 also participates in these stacking interactions and serves as the boundary between the ligand binding pocket and the sodium binding pocket. [1]
Na+ Binding Pocket
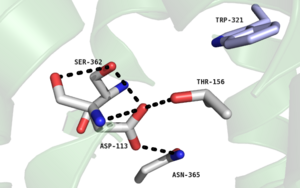
Figure 2. Residues of collapsed sodium binding pocket. Trp321 (blue) sets the top of the pocket, where Ser362, Asp113, Thr156 and Asn365 (gray) are involved in hydrogen bonding interactions preventing the coordination of a Na
+ ion.
, which is positioned at the bottom of the , sets the top of the . The Na+ ion binding pocket acts as a negative allosteric site for G protein activity [1]. When Na+ enters the Na+ ion binding pocket, it coordinates with Asp95, Gln131, Ser135, and Asp113, decreasing the signaling activity of NTSR1 [1]. When NTSR1 is in its active state, the Na+ ion binding pocket is collapsed, preventing the regulations of protein activity through a Na+ ion, as the Na+ ion is unable to be coordinated by a salt bridge to Asp113 (Figure 2). The side chain atoms of Asp113 form a hydrogen bond network with Thr156, Ser361, Ser362, and Gln365 instead, which prevents the coordination of a Na+ ion[1] (Figure 2).
Activation of NTSR1
Since wild type NTSR1 was unstable in detergent solution, six residues in the protein were mutated for stabilization. [2] [1]
Active-Like State
The, six amino acids that were mutated for thermostabilization [2] were A86L, E166A, G215A, L310A, F358A, and V360A. This protein was found to have NTS affinity similar to that of wild tpye NTSR1, and was named . Along with this, the Na+ ion binding pocket was found to be collapsed in this protein. However, researchers found that NTSR1-GW5 did not have G-protein activity [2].
Active State
After determining as only active-like, research was conducted to determine the structure of NTSR1 with catalytic nucleotide exchange. In order to do so, three of the six mutations were reverted back [1], and these three residues were selected on the basis of their location. The reversion of E166A, L310A, and F358A led to NTSR1 with G-protein activity at almost wild-type level. This protein was named, , and indicated that the amino acid residues E166, L310, and F358 play significant roles in the activity of NTSR1 [1].
Leu310
is crucial for interactions with the G alpha subunit by interacting with Arg167 in the D/ERY motif [1] . When Leu310 was substituted with alanine, Arg167 was able to form a stabilizing hydrogen bonding network with Asn257, Ser164 and Gly306, which oriented Arg167 in a position that was unfavorable for contacting the G alpha subunit. When residue 310 was converted back to leucine, this hydrogen-bonding network was sterically unfavorable (Figure 3) and Arg167 interacted with the G alpha subunit leading to the transduction of several different signals involved in leptin signlaing, dopamine regulation, etc. [1] [3][4][5]
.
Phe358
When this residue was mutated to an alanine [1] the of the ligand binding pocket were interrupted,resulting in a lack of G-Protein activity in NTSR1.[1].This supported the role of Phe358 as stated in the hydrophobic binding pocket section of this page.
Glu166
Although the role of in G-protein activity is not quite as clear as it is for or , substituting this residue for an alanine significantly reduced catalytic nucleotide exchange [1]. Glu166 is part of a D/ERY motif that is highly conserved in class A GPCRs and includes Arg167 and Tyr168. To determine a role for Glu166, other class A GPCRs were structurally analyzed. Researchers [1] hypothesize that E166 in NTSR1 interacts with Val102, Thr101,and weakly interacts with His105 to stabilize the G protein. Researchers [1] have also noted the possibility of an important connection between the D/ERY motif and intracellular loop 2, which plays a role in the dissociation of the receptor-G protein complex with GTP present. M181 is believed to link the D/ERY motif and ICL2. [1]
Biological Relevance
Neurotensin
is a 13 amino acid peptide that is found in both nervous and peripheral tissues. It functions as a hormone and a neurotransmitter by activating the G-protein coupled receptor NTSR1[1]
Leptin Research
NTSR1 deficient mice were not able to receive a satiety signal.[3]The mice continued to eat when food was present, leading to significant weight gain. This relationship between satiety and NTS is related to the involvement of NTSR1 in the signaling pathway for Leptin and therefore food intake. Without sufficient NTSR1 this pathway is interrupted. [3]
Cancer Studies
Some tumor cells can secrete and express Neurotensin and Neurotensin receptors themselves suggesting that Neurotensin autocrine, endocrine and paracrine regulation are possible. This leads to aggressive growth and tumor development. Injecting animals with Neurotensin increased tumor growth and size, while injecting them with Neurotensin antagonist decreased tumor growth [4].Neurotensin regulation may be used in future cancer treatments.
Dopamine Regulation
The dopamine hypothesis states that hyperdopamine levels may lead to schizophrenic symptoms. NTSR1 caused a blockade which inhibited firing in dopaminergic cells suggesting that NTSR1 could be used in schizophrenia treatment. The secondary effects were too extreme and the trial was discontinued.NTSR1 research focuses on dopamine regulation for treating schizophrenia.[5]


