We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1176
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
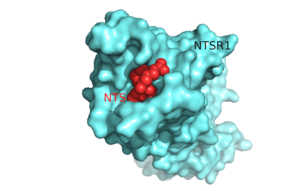
On the extracellular side of the protein is the | On the extracellular side of the protein is the | ||
<scene name='72/721547/Hydrophobic_binding_pocket/5'>hydrophobic binding pocket</scene>. <ref name="SONT"/> | <scene name='72/721547/Hydrophobic_binding_pocket/5'>hydrophobic binding pocket</scene>. <ref name="SONT"/> | ||
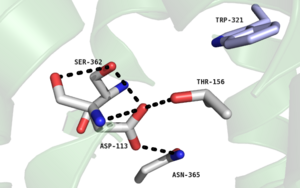
| - | One key residue in this pocket is a Phenylalanine at position 358, which takes part in a network of hydrophobic stacking interactions | + | One key residue in this pocket is a Phenylalanine at position 358, which takes part in a network of hydrophobic stacking interactions<ref name="SPGP"/>. These interactions stabilize the Trp321 and Tyr324 residues allowing Tyr324 to interact with the '''[https://en.wikipedia.org/wiki/C-terminus C-terminal]''' |
<scene name='72/721547/Hydrophobic_binding_pocket/6'>Leu13 residue of the NTS ligand</scene> | <scene name='72/721547/Hydrophobic_binding_pocket/6'>Leu13 residue of the NTS ligand</scene> | ||
via '''[https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals interactions]''' .<ref name="SONT"/><ref name="SPGP"/> | via '''[https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals interactions]''' .<ref name="SONT"/><ref name="SPGP"/> | ||
| Line 20: | Line 20: | ||
==Activation of NTSR1== | ==Activation of NTSR1== | ||
| - | Since wild type NTSR1 was unstable in detergent solution for imaging, six residues in the protein were mutated for stabilization. | + | Since wild type NTSR1 was unstable in detergent solution for imaging, six residues in the protein were mutated for stabilization.<ref name="SONT"/> <ref name="SPGP"/> |
===Active-Like State=== | ===Active-Like State=== | ||
Revision as of 02:52, 22 April 2016
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 Krumm BE, White JF, Shah P, Grisshammer R. Structural prerequisites for G-protein activation by the neurotensin receptor. Nat Commun. 2015 Jul 24;6:7895. doi: 10.1038/ncomms8895. PMID:26205105 doi:http://dx.doi.org/10.1038/ncomms8895
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012 Oct 25;490(7421):508-13. doi: 10.1038/nature11558. Epub 2012 Oct 10. PMID:23051748 doi:http://dx.doi.org/10.1038/nature11558
- ↑ 3.0 3.1 3.2 3.3 Liang Y, Boules M, Li Z, Williams K, Miura T, Oliveros A, Richelson E. Hyperactivity of the dopaminergic system in NTS1 and NTS2 null mice. Neuropharmacology. 2010 Jun;58(8):1199-205. doi:, 10.1016/j.neuropharm.2010.02.015. Epub 2010 Mar 6. PMID:20211191 doi:http://dx.doi.org/10.1016/j.neuropharm.2010.02.015
- ↑ 4.0 4.1 4.2 Carraway RE, Plona AM. Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides. 2006 Oct;27(10):2445-60. Epub 2006 Aug 2. PMID:16887236 doi:http://dx.doi.org/10.1016/j.peptides.2006.04.030
- ↑ 5.0 5.1 5.2 Griebel G, Holsboer F. Neuropeptide receptor ligands as drugs for psychiatric diseases: the end of the beginning? Nat Rev Drug Discov. 2012 May 18;11(6):462-78. doi: 10.1038/nrd3702. PMID:22596253 doi:http://dx.doi.org/10.1038/nrd3702