Introduction
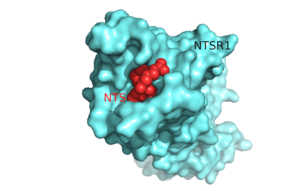
Figure 1.Top view of NTSR1 protein (blue) interacting with its ligand, NTS(red).
Neurotensin receptor 1 (NTSR1) is a G-protein coupled receptor (GPCR). GPCRs are a class of proteins with an extracellular binding domain and 7 transmembrane helices that assist in propagating a cellular response[1]. This is accomplished by the binding of ligands to the GPCR outside the cell, causing a conformational change and activating a signal transduction pathway via second messengers such as cyclic AMP, inositol triphosphate, and diacylglycerol.[1] The ligand for NTSR1 is the 13 amino acid peptide, neurotensin (NTS)[2], and the majority of the effects of NTS are mediated through NTSR1[2]. NTS has a variety of biological activities including a role in the leptin signaling pathways [3], tumor growth [4], and dopamine regulation [5]. Recently NTSR1 was crystallized bound with the C-terminus of its tridecapeptide ligand, . The shortened ligand was used because it has a higher potency and efficacy than its full-length counterpart[2]. Class A GPCRs bind their ligands within the transmembrane core in a ligand binding pocket. The in NTSR1 is located at the top of the protein (Figure 1). NTSR1 also contains an allosteric , which is located directly beneath the ligand binding pocket and the two pockets are separated by the residue [1]. NTSR1 has been mutated to exist in both and states. This has led to a greater understanding of the structure of NTSR1 and how the structure influences its function.
Structure
Ligand Binding Pocket
On the extracellular side of the protein is the
. [2]
One key residue in this pocket is a Phenylalanine at position 358, which takes part in a network of hydrophobic stacking interactions[1]. These interactions stabilize the Trp321 and Tyr324 residues allowing Tyr324 to interact with the C-terminal
via Van der Waals interactions .[2][1]
Without the hydrophobic stacking interactions that are facilitated by the Phe358, this binding interaction would be destabilized. Trp321 also participates in these stacking interactions and serves as the boundary between the ligand binding pocket and the Na+ binding pocket.[1]
Na+ Binding Pocket
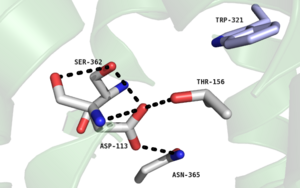
Figure 2. Residues of the collapsed sodium binding pocket. Trp321 (blue) sets the top of the pocket, where Ser362, Asp113, Thr156 and Asn365 (gray) are involved in hydrogen bonding interactions preventing the coordination of a Na
+ ion.
, which is positioned at the bottom of the , sets the top of the . The Na+ ion binding pocket acts as a negative allosteric site for G protein activity [1]. When Na+ enters the Na+ ion binding pocket, it coordinates with Asp95, Gln131, Ser135, and Asp113, decreasing the signaling activity of NTSR1 [1]. When NTSR1 is in its active state, the Na+ ion binding pocket is collapsed. This prevents the regulation of protein activity through a Na+ ion, as the Na+ ion is unable to coordinate via a salt bridge to Asp113 (Figure 2). The side chain atoms of Asp113 form a hydrogen bond network with Thr156, Ser361, Ser362, and Gln365 instead, which prevents the coordination of a Na+ ion[1] (Figure 2).
Activation of NTSR1
Since wild type NTSR1 was unstable in detergent solution for imaging, six residues in the protein were mutated for stabilization.[2] [1]
Active-Like State
The six amino acid mutations for thermostabilization [2] were Ala86Leu, Glu166Ala, Gly215Ala, Leu310Ala, Phe358Ala, and Val360Ala. This protein was found to have NTS affinity similar to that of wild tpye NTSR1, and was named . Along with this, the Na+ ion binding pocket was collapsed in this protein. However, NTSR1-GW5 did not have G-protein activity [2].
Active State
After determining the original structural state, , as only active-like, the structure of NTSR1 was determined in an active state.[1] By reverting back three of the original six mutations from the active-like structure on the basis of their location[1], NTSR1 gained near wild-type activity.[1] The three reversions were Asp166, Leu310, and Phe358, and this protein was named . The revival of activity in NTSR1 indicated that the reverted amino acid residues (Asp166, Leu310, and Phe358) play significant roles in G-protein activity.[1]
Leu310
is crucial for interactions with the G alpha subunit by positioning Arg167 in the conserved [1]. When Leu310 was substituted with alanine, Arg167 was able to form a stabilizing hydrogen bonding network with Asn257, Ser164 and Gly306, which oriented Arg167 in a position that was unfavorable for contacting the G alpha subunit. When residue 310 was converted back to leucine, this hydrogen bonding network was sterically unfavorable and Arg167 interacted with the G alpha subunit[1] leading to the transduction of several different signals involved in dopamine regulation[5], leptin signlaing[3], and tumor growth[4].
Phe358
When this residue was mutated to an alanine [1] the of the ligand binding pocket were interrupted,resulting in a lack of G-Protein activity in NTSR1.[1].This supported the role of Phe358 as stated in the hydrophobic binding pocket section of this page.
Glu166
Although the role of in G-protein activity is not quite as clear as it is for or , substituting this residue for an alanine significantly reduced catalytic G-protein activity [1]. Glu166 is part of a that is highly conserved in class A GPCRs and includes Arg167 and Tyr168. It's hypothesized [1] that Glu166 interacts with Val102, Thr101,and His105 to stabilize the G protein. An important connection between the D/ERY motif and intracellular loop 2 via M181, has also been hypothesized.[1]. ICL2 plays a role in the dissociation of the receptor-G protein complex with GTP present.[1]
Biological Relevance
Neurotensin
is a 13 amino acid peptide that is found in both nervous and peripheral tissues [1]. It functions as a hormone and a neurotransmitter by activating the G-protein coupled receptor, NTSR1[1].
Leptin Research
NTSR1 deficient mice were not able to receive a satiety signal from Leptin[3]. The mice continued to eat when food was present, leading to significant weight gain. With an NTSR1 deficiency, NTS does not bind efficiently to NTSR1, and the leptin signaling pathway is interrupted [3].
Cancer Studies
Some tumor cells can secrete and express NTS and NTS receptors themselves suggesting that NTS autocrine, endocrine and paracrine regulation are possible. This leads to aggressive growth and tumor development. Injecting animals with NTS increased tumor growth and size, while injecting them with NTS antagonist had the opposite effect [4]. NTS regulation may be used in future cancer treatments.
Dopamine Regulation
The dopamine hypothesis states that hyperdopamine levels may lead to schizophrenic symptoms. NTSR1 causes a blockade which inhibits firing in dopaminergic cells suggesting that NTSR1 could be used in schizophrenia treatment. However, this led to extreme secondary effects and was discontinued. Despite this, research on NTSR1 as a treatment for schizophrenia persists[5].
Neurotensin Receptor (NTSR1)
Introduction
The neurotensin receptor (NTSR1) belongs to the superfamily of proteins known as G protein-coupled receptors (GPCRs) and responds to the 13 amino acid hormone neurotensin (NTS). Currently around 800 G protein-coupled receptors have been identified and are hypothesized to be responsible for roughly 80% of signal transduction.[6] GPCRs are involved in a vast array of physiological processes within the body that range from interactions with dopamine to effects on secretion of bile in the intestines.[7] [8] Due to the vast array of functions that these proteins serve and their high abundance within the body, these proteins have become major drug targets.[9]
A critical topic in the understanding of GPCRs is the transition from the inactive to active state. This transition is responsible for the transduction of a signal from the extracellular to the intracellular space. The transition occurs when a ligand, NTS in the case of NTSR1, binds to the receptor causing a conformational change in the protein that leads to the activation of the intracellular G protein. Currently no crystal structures of the receptor in its unbound, inactive form exist making the transition difficult to study. [10] NTSR1 can be seen in blue and the ligand NTS can be seen in green.
Neurotensin
Neurotensin (NTS) is a 13-amino acid peptide originally isolated from bovine hypothalamus. [11] NTS fulfills the roles of both a neurotransmitter and a neuromodulator in the nervous system and a hormone in the periphery nervous system. NTS is a neuromodulator of dopamine transmission and of anterior pituitary hormone secretion. [12] In the periphery of the digestive tract and cardiovascular system, NTS is a paracrine and endocrine modulator.[13] Finally, NTS serves as a growth factor for many normal and cancerous cell types. [14]
Only the C-terminal tail of NTS, amino acids 8-13, were resolved in the .(PDB code:4GRV) Amino acids 8-13 are only resolved because these are the amino acids that interact with NTRS1 to produce the conformational change that is responsible for G-protein activation. The peptide is colored by element: carbon, oxygen, nitrogen.
Structure
Overall Structure
Figure 1: Neurotensin Incorporation in Membrane. This image depicts the spanning of the membrane made by NTSR1 and illustrates the need for transduction from its extracellular binding site to the intracellular region. (PubMed).
Like other G protein-coupled receptors, the neurotensin receptor is composed of 3 distinct regions. An where neurotensin binds and causes a conformational change of the protein. A region containing (PDB code:4GRV) that transduce the signal from the extracellular side of the cell membrane to the intracellular side. Lastly, an intracellular region, that when activated by a conformational change in the protein, activates a G-protein associated with this receptor. Currently no crystal structures of the inactive form of the neurotensin receptor are available. Without a representation of the inactive form, the conformational changes caused by agonist binding are still not completely known.
Neurotensin Binding Site
Binding of NTS to the binding site on NTSR1 is enriched by (PDB code:4GRV)between the positive NTS arginine side chains and the electronegative pocket. The protein is colored by charge: negative and positive. Two of NTS's arginine residues are colored blue. In addition, the C-terminus of NTS forms a (PDB code:4GRV) with Arg328 of NTSR1. Three hydrogen bonds are made between the side chains of NTS and the receptor while most of the interactions are a result of Van der Waals interactions. The binding pocket is partially capped by a at the proximal end of the receptor's N-terminus.[10] The interactions in the binding site cause a wide spread conformational change in the receptor leading to the receptor to adopt an active conformation. This active conformation allows for the activation of the associated intracellular G-protein.
Hydrophobic Stacking
A major player in the transduction of the extracellular signal to the intracellular G-protein is the (PDB code:4GRV)that links the bound hormone with the hydrophobic core of the neurotensin receptor. The carboxylate of Leu13 of NTS forms a hydrogen bond network with R327, R328, and Y324. The Tyr324, in turn, is brought into an orientation to make the formation of a (PDB Code:4XEE) network between F358, W321, A157, and F317 possible.[15] The effects that this network has on the activation of the intracellular G-protein was examined by the mutagenesis of amino acids that disrupted the formation of this network. Mutagenesis of , , , , , and (PDB code:4GRV) showed that when this interaction was disrupted, the receptor no longer was able to activate the G-protein. [15] This discovery lead to the conclusion that the conformational changes caused by this stacking allows for the signal to be moved from the extracellular binding site through the transmembrane helices of the receptor to the intracellular region activating the G-protein.
Sodium Binding Pocket
Figure 2: Closed form of sodium binding pocket that caps the entrance of sodium into the top of the binding pocket. (PDB Code:
4XEE)
Figure 3: Open form of sodium binding pocket that does not cap the entrance of sodium into the top of the binding pocket. (PDB code:
4GRV)
Conserved across all class A GPCRs, a (PDB code:
4GRV) is seen in the middle of TM2 helix. The sodium ion is coordinated with a highly conserved Asp113 and four other oxygen contacts from a combination of water molecules. For G-protein activation to be possible, a hydrogen bond coordination with T156, S362, and N365 of the NPxxY
motif must occur. Trp321 helps to maintain the active conformation of the receptor by occluding the top of the binding pocket using
Van der Waals interactions (Figure 2). This occlusion stops sodium ions from entering the top of the binding pocket and helps NTSR1 remain in its active conformation. The conformation of the binding pocket where Trp321 does not occlude the top can be seen when mutations to A86L, G215A, and V360A are present (Figure 3). This form of the receptor would allow more sodium into the binding pocket and therefor stabilize the inactive receptor form.
[16]
Allosteric Effects
Sodium ions are a negative allosteric inhibitor to the binding of the neurotensin agonist to the binding site on the neurotensin receptor. Sodium's binding causes for the receptor to favor its inactive state by disrupting the hydrogen bond network between nearby amino acids. This disruption in the hydrogen bond network causes the pocket to be in its uncollapsed form. Asp113 of the highly conserved D/RY motif and Asn365 of the highly conserved NPxxY motif form a substantial hydrogen bonding network with T156 and S362.[15] This hydrogen bonding network prevents the incorporation of the sodium ion by collapsing upon itself and filling the sodium binding pocket. Trp321 also works to inhibit the incorporation of the sodium ion by capping off the sodium binding pocket to not allow sodium to enter from the top. Trp321 uses Van der Waals interactions to place it in the conformation necessary to block sodium from entering the site. By not allowing for sodium to enter this binding site, the receptor is able to conform to its active state and activate the G-protein that is associated with it.
Clinical Relevance
NTSR1 is commonly expressed in various invasive
cancer cell lines making it a promising cancer drug target. It is prevalent in
colon cancer adenocarcinoma, but is not found in adult colon cell types.
[17] NTSR1 is also found in aggressive
prostate cancer cells, but not
epithelial prostate cells. In prostate cancer cells, binding of NTS results in
mitogen-activated protein kinase (PKB),
phosphoinositide-3 kinase (PI-3K),
epidermal growth factor receptor (EGFR),
SRC, and
STAT5 phosphorylation.
[17] These all result in increased DNA synthesis,
cell proliferation, and survival. Inhibition of NTSR1 and its downstream signaling represents a target for
radiotherapy, which uses radiation to target malignant cells.
Figure 4: Meclinerant: An inhibitor of NTSR1 found to enhance selectivity of radiotherapy in cancer treatment (PubMed).
NTSR1 can be inhibited by agonist
meclinertant which inhibits proliferation and prosurvival of cancer cells. Combination treatment of radiation and meclinerant provides selective treatment of cancer cells over normal cells, indicating the need for clinical trials of this approach.
[18]
StructureSection | PDB=4GRV2 | Size =400 | Side=right | scene ='72/727765/Overall_structure/2' | caption='Neurotensin G-Protein Coupled Receptor (PDB Codes 4GRV and 4XEE)'






