User:Madelyn Kasprzak/Sandbox 6
From Proteopedia
(Difference between revisions)
m (Added short description to structural highlights section) |
|||
| Line 6: | Line 6: | ||
== Structure == | == Structure == | ||
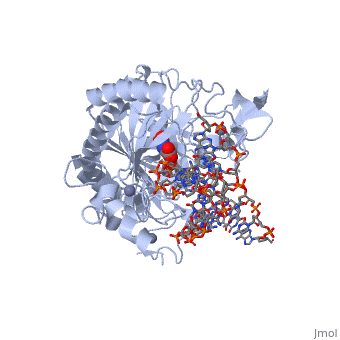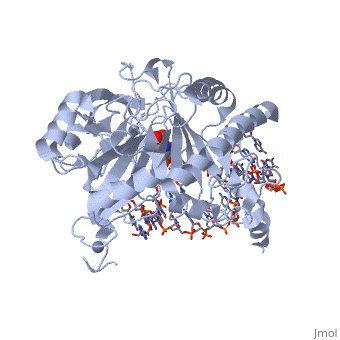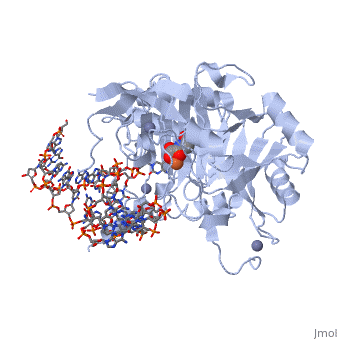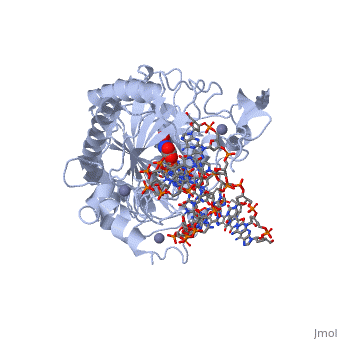
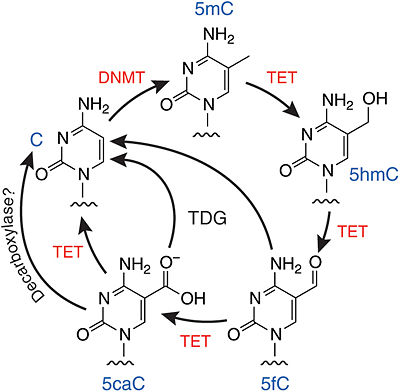
| - | The TET enzymes have a cysteine-rich region closely followed by a double stranded beta-helix (DSBH) domain near their C-terminus.<ref name='Kinney et al.'>DOI 10.1007/978-1-4419-9967-2_3</ref> The DSBH domain contains three Fe<sup>2+</sup> binding sites and an α-ketoglutarate binding site.<ref name='Kinney et al.' /> | + | The TET enzymes have a cysteine-rich region closely followed by a double stranded beta-helix (DSBH) domain near their C-terminus.<ref name='Kinney et al.'>DOI 10.1007/978-1-4419-9967-2_3</ref> The DSBH domain contains three Fe<sup>2+</sup> binding sites and an α-ketoglutarate binding site.<ref name='Kinney et al.' /> For all α-ketoglutarate oxygenases, including TET enzymes, the DSBH domain and the preceding cysteine-rich region, perform the main catalytic activity for these enzymes. |
In addition, TET1 has a CXXC-type zinc finger domain near the N-terminus. However, the TET1 CXXC domain lacks the conserved lysine-phenylalanine-glycine-glycine (KFGG) motif commonly seen within the CXXC domains of other DNA binding proteins, such as DNA methyltransferase-1 (DNMT1). A study conducted by Frauer et al. in 2011 showed that the isolated CXXC domain of TET1 has no DNA binding activity, which agrees with the evidence suggesting that the KFGG motif increases affinity for unmethylated DNA.<ref name='Frauer et al.'>DOI 10.1371/journal.pone.0016627</ref> Frauer et al. also speculated that the CXXC domain of TET1 may be involved with protein-protein interactions instead of DNA binding.<ref name='Frauer et al.' /> | In addition, TET1 has a CXXC-type zinc finger domain near the N-terminus. However, the TET1 CXXC domain lacks the conserved lysine-phenylalanine-glycine-glycine (KFGG) motif commonly seen within the CXXC domains of other DNA binding proteins, such as DNA methyltransferase-1 (DNMT1). A study conducted by Frauer et al. in 2011 showed that the isolated CXXC domain of TET1 has no DNA binding activity, which agrees with the evidence suggesting that the KFGG motif increases affinity for unmethylated DNA.<ref name='Frauer et al.'>DOI 10.1371/journal.pone.0016627</ref> Frauer et al. also speculated that the CXXC domain of TET1 may be involved with protein-protein interactions instead of DNA binding.<ref name='Frauer et al.' /> | ||
Revision as of 17:17, 27 April 2016
TET Enzymes
TET enzymes are a family of dioxygenases that are involved in the process of oxidizing methylated cytosine. Members of this family include ten-eleven translocation methylcytosine dioxygenase 1 (), methylcytosine dioxygenase , and methylcytosine dioxygenase . The gene for the first of these proteins, TET1, was identified when it was determined to be fused to the Mixed Lineage Leukemia (MLL) gene as a result of a translocation event that occurred between chromosomes ten and eleven (hence the name). [1]
| |||||||||||
References
- ↑ Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003 Mar;17(3):637-41. PMID:12646957 doi:http://dx.doi.org/10.1038/sj.leu.2402834
- ↑ 2.0 2.1 Kinney SR, Pradhan S. Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer. Adv Exp Med Biol. 2013;754:57-79. doi: 10.1007/978-1-4419-9967-2_3. PMID:22956496 doi:http://dx.doi.org/10.1007/978-1-4419-9967-2_3
- ↑ 3.0 3.1 Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, Wang M, Qin W, Soding J, Spada F, Leonhardt H. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011 Feb 2;6(2):e16627. doi: 10.1371/journal.pone.0016627. PMID:21311766 doi:http://dx.doi.org/10.1371/journal.pone.0016627
- ↑ 4.0 4.1 4.2 He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011 Sep 2;333(6047):1303-7. doi: 10.1126/science.1210944. Epub 2011 Aug, 4. PMID:21817016 doi:http://dx.doi.org/10.1126/science.1210944
- ↑ 5.0 5.1 Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013 Oct 24;502(7472):472-9. doi: 10.1038/nature12750. PMID:24153300 doi:http://dx.doi.org/10.1038/nature12750
- ↑ Huang Y, Rao A. New functions for DNA modifications by TET-JBP. Nat Struct Mol Biol. 2012 Nov;19(11):1061-4. doi: 10.1038/nsmb.2437. PMID:23132381 doi:http://dx.doi.org/10.1038/nsmb.2437
- ↑ Takai H, Masuda K, Sato T, Sakaguchi Y, Suzuki T, Suzuki T, Koyama-Nasu R, Nasu-Nishimura Y, Katou Y, Ogawa H, Morishita Y, Kozuka-Hata H, Oyama M, Todo T, Ino Y, Mukasa A, Saito N, Toyoshima C, Shirahige K, Akiyama T. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014 Oct 9;9(1):48-60. doi: 10.1016/j.celrep.2014.08.071. Epub 2014 Oct, 2. PMID:25284789 doi:http://dx.doi.org/10.1016/j.celrep.2014.08.071
- ↑ 8.0 8.1 Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009 May 28;360(22):2289-301. doi: 10.1056/NEJMoa0810069. PMID:19474426 doi:http://dx.doi.org/10.1056/NEJMoa0810069
- ↑ Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009 Jul;41(7):838-42. doi: 10.1038/ng.391. Epub 2009 May 31. PMID:19483684 doi:http://dx.doi.org/10.1038/ng.391
- ↑ Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002 Jul 15;62(14):4075-80. PMID:12124344
- ↑ Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003 Mar;17(3):637-41. PMID:12646957 doi:http://dx.doi.org/10.1038/sj.leu.2402834