Nos1
From Proteopedia
| Line 4: | Line 4: | ||
== Location == | == Location == | ||
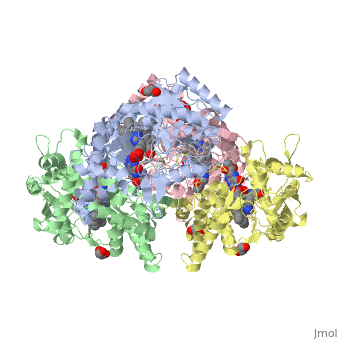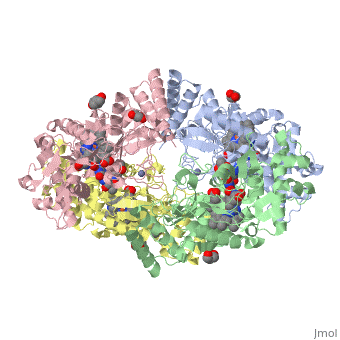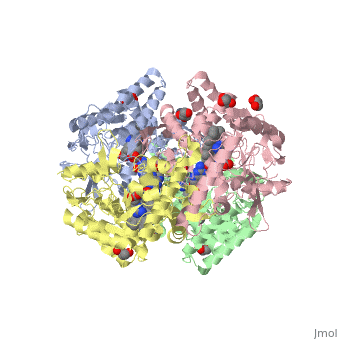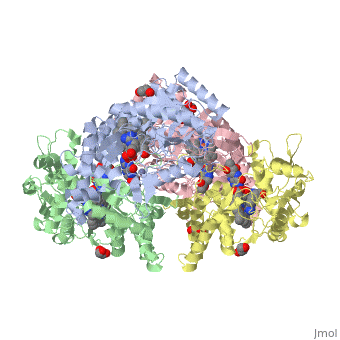
| - | The gene NOS1, expressed by humans, encodes for an enzyme in the nitric oxide synthase family. Nitric oxide synthases consume L-arginine and molecular oxygen to form the free radical nitric oxide, which | + | The gene NOS1, expressed by humans, encodes for an enzyme in the nitric oxide synthase family. Nitric oxide synthases consume L-arginine and molecular oxygen to form the free radical nitric oxide, which acts as a signaling molecule in a wide range of molecular and biological processes. <ref>Hou, YC; Janczuk, A; Wang, PG (1999). "Current trends in the development of nitric oxide donors". Current pharmaceutical design 5 (6): 417–41.</ref> The reaction catalyzed by nitric oxide synthase is shown below. <ref>Knowles RG, Moncada S (March 1994). "Nitric oxide synthases in mammals". Biochem. J. 298 (2): 249–58. PMC 1137932. PMID 7510950.</ref> |
| - | 2 L-arginine + 3 NADPH + 4 O(2) = 2 L- citrulline + 2 nitric oxide + 3 NADP(+) + 4 H(2)O | + | 2 L-arginine + 3 NADPH + 4 O(2) = 2 L- citrulline + 2 nitric oxide + 3 NADP(+) + 4 H(2)O |
Nitric oxide is used in a wide variety of processes. A few examples of molecular processes include heme binding, NADP binding, calcium signaling, and oxidation-reduction reactions. Biological examples include regulation of cardiac muscle contraction, blood coagulation, aging, and regulation of neurogenesis <ref>UniProt Consortium 2009, ‘UniProtKB - P29475 (NOS1_HUMAN),’ UniProtKB Protein Knowledgebase</ref>. There are three known isoforms known for NOS1 in humans produced by alternative splicing, as well as four natural transcript variants for NOS1. The alternatively-spliced isoforms are reffered to as neuronal NOS (nNOS or NOS1), endothelial NOS (eNOS or NOS3), and cytokine-inducible NOS (iNOS or NOS2). Moderate expression of NOS1 has been observed in skeletal muscle, brain, testicular, lung, and kidney tissue. Low expression has been observed in heart, adrenal gland, and retinal tissues <ref>Ward ME, Toporsian M, Scott JA, et al. Hypoxia induces a functionally significant and translationally efficient neuronal NO synthase mRNA variant. Journal of Clinical Investigation. 2005;115(11):3128-3139. doi:10.1172/JCI20806.</ref>. | Nitric oxide is used in a wide variety of processes. A few examples of molecular processes include heme binding, NADP binding, calcium signaling, and oxidation-reduction reactions. Biological examples include regulation of cardiac muscle contraction, blood coagulation, aging, and regulation of neurogenesis <ref>UniProt Consortium 2009, ‘UniProtKB - P29475 (NOS1_HUMAN),’ UniProtKB Protein Knowledgebase</ref>. There are three known isoforms known for NOS1 in humans produced by alternative splicing, as well as four natural transcript variants for NOS1. The alternatively-spliced isoforms are reffered to as neuronal NOS (nNOS or NOS1), endothelial NOS (eNOS or NOS3), and cytokine-inducible NOS (iNOS or NOS2). Moderate expression of NOS1 has been observed in skeletal muscle, brain, testicular, lung, and kidney tissue. Low expression has been observed in heart, adrenal gland, and retinal tissues <ref>Ward ME, Toporsian M, Scott JA, et al. Hypoxia induces a functionally significant and translationally efficient neuronal NO synthase mRNA variant. Journal of Clinical Investigation. 2005;115(11):3128-3139. doi:10.1172/JCI20806.</ref>. | ||
Revision as of 03:01, 28 April 2016
| |||||||||||
References
Shinkai, T., Ohmori, O., Hori, H., and Nakamura, J. (2002) Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Molecular Psychiatry. 7, 560-563. doi:10.1038/sj.mp.4001041 Galimberti, D., Scarpini, E., Venturelli, E., Strobel, A., Herterich, S., Fenogolio, C., Guidi, I., Scalabrini, D., Cortini, F., Bresolin, N., Lesch, K., and Reif, A. (2008) Association of a NOS1 promoter repeat with Alzheimer’s disease. Neurobiology of Aging. 29, 1359-1365. doi:10.1016/j.neurobiolaging.2007.03.003 Rife, T., Rasoul, B., Pullen, N., Mitchell, D., Grathwol, K., and Kurth, J. (2009) The effect of a promoter polymorphism on transcription of nitric oxide synthase 1 and its relevance to Parkinson’s disease. Journal of Neuroscience Research. 87, 2319-2325. doi:10.1005/jnr.22045
- ↑ Hou, YC; Janczuk, A; Wang, PG (1999). "Current trends in the development of nitric oxide donors". Current pharmaceutical design 5 (6): 417–41.
- ↑ Knowles RG, Moncada S (March 1994). "Nitric oxide synthases in mammals". Biochem. J. 298 (2): 249–58. PMC 1137932. PMID 7510950.
- ↑ UniProt Consortium 2009, ‘UniProtKB - P29475 (NOS1_HUMAN),’ UniProtKB Protein Knowledgebase
- ↑ Ward ME, Toporsian M, Scott JA, et al. Hypoxia induces a functionally significant and translationally efficient neuronal NO synthase mRNA variant. Journal of Clinical Investigation. 2005;115(11):3128-3139. doi:10.1172/JCI20806.
- ↑ Juliane Kopf, Martin Schecklmann, Tim Hahn, Thomas Dresler, Alica C. Dieler, Martin J. Herrmann, Andreas J. Fallgatter, Andreas Reif. (2011) NOS1 ex1f-VNTR polymorphism influences prefrontal brain oxygenation during a working memory task, NeuroImage, Volume 57, (Issue 4),1617-1623, Ihttp://dx.doi.org/10.1016/
- ↑ Freudenberg, F., Alttoa, A. & Reif, A. (2015) Neuronal nitric oxide synthase (NOS1) and its adaptor (NOS1AP) act as a genetic risk factors for psychiatric. Genes Brain Behav 14, 47–64.
- ↑ Zhang, Y. H., Jin, C. Z., Jang, J. H., & Wang, Y. (2014). Molecular mechanisms of neuronal nitric oxide synthase in cardiac function and pathophysiology. The Journal of Physiology, 592(Pt 15), 3189–3200. http://doi.org/10.1113/jphysiol.2013.270306]
- ↑ Zhang, Y. H., Jin, C. Z., Jang, J. H., & Wang, Y. (2014). Molecular mechanisms of neuronal nitric oxide synthase in cardiac function and pathophysiology. The Journal of Physiology, 592(Pt 15), 3189–3200. http://doi.org/10.1113/jphysiol.2013.270306]
Li H, Jamal J, Plaza C, et al. Structures of human constitutive nitric oxide synthases. Acta Crystallographica Section D: Biological Crystallography. 2014;70(Pt 10):2667-2674.