Structural highlights
1xpb is a 1 chain structure with sequence from Escherichia coli. Full crystallographic information is available from OCA. For a guided tour on the structure components use FirstGlance.
|
| Ligands: | |
| Activity: | Beta-lactamase, with EC number 3.5.2.6 |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum |
| Related: | 1axb, 1bt5, 1btl, 1ck3, 1erm, 1ero, 1erq, 1esu, 1fqg, 1jtd, 1jtg, 1jvj, 1jwp, 1jwv, 1jwz, 1lhy, 1li0, 1li9, 1m40, 1nxy, 1ny0, 1nym, 1nyy, 1pzo, 1pzp, 1s0w, 1tem, 1xxm, 1yt4, 1zg4, 1zg6, 2v1z |
Background and History
Antibiotics have long been the primary line of defense against many infections. The advent of penicillin in the 1930s brought an end to an era of uncertainty: diseases once considered fatal became treatable. Unfortunately, natural selection stops for no species. Resistance to some form of antibiotic now has become the standard for many infections.[1] Moreover, their liberal usage has resulted in “superbugs”, which are resistant to multiple antibiotics.[2] These strains are capable of secreting enzymes capable of deactivating the antibiotic or modifying their cell walls to render the antibiotic ineffective.
Many antibiotics such as penicillin, their derivatives (penams), and cephalosporins (cephams) possess a β-lactam ring (Figure 1).
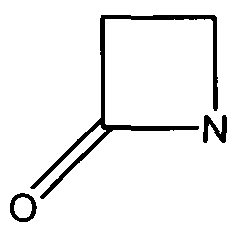
Figure 1. The Beta-Lactam ring is shown above. Beta-lactamases function in hydrolyzing the amide bond within the ring, rendering the antibiotic ineffective. (Image Credit: Wikipedia)
β-Lactamases function by hydrolyzing the β-lactam ring within the antibiotic (Figure 2). This prevents the interaction between the cell wall and the antibiotic.
Figure 2. Bacteria are capable of becoming antibiotic resistant by catalyzing the hydrolysis of the β-lactam ring.
The TEM-1 subclass is one of the most common of all β-Lactamases.[3] Their prevalence began when bacteria started exhibiting penicillin resistance on a mass scale. Since the TEM-1 subclass has been prevalent for many years, many inhibitors have either been discovered or synthesized to prevent their catalytic function.
Function
The β-Lactamases has two domains- an alpha helix and a beta sheet of five antiparallel strands, which surround the alpha helix. The catalytic region is known as the oxyanion pocket that occurs between the N-terminus of alpha helix H2 and B3 beta sheet edge. The first step is the acylation of Ser70, one of the catalytic sites. This forms a high-energy acyl-enzyme intermediate, which is then deacylated. This is when the acyl-enzyme is hydrolyzed. The β-Lactam compound has been split and released. The rate determining step is either the acylation or deacylation depending on the antibiotic. However, the opening of the β-Lactam active site increases its chance of inhibition. One of the ways to inhibit β-Lactamases is point mutations in the catalytic region that affect the specificity and catalysis of β-Lactamases enzymes, hindering their activity [4].
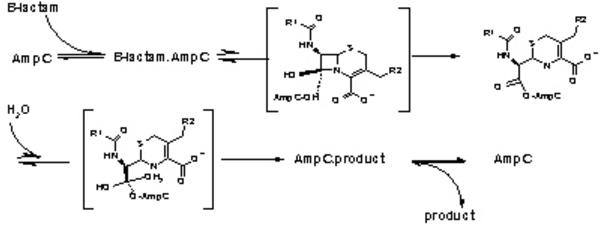
Figure 3. The hydrolysis of β-Lactamases where AmpC is the enzyme. There are the three main steps shown with their respective high energy intermediates. (Image Credit: Shoichet Lab in UCSF)
Relevance
TEM-1 class antibiotic proteins are probably the most well-known antibiotic resistant protein in the world. Nowadays antibiotic resistance is becoming an ever increasing problem in the medical community. The lives of antibiotics are decreasing as resistant strands of bacteria become more prevalent. In the year 2003, 65% of antibiotics were β-lactamase derivatives [5]. Resistance to these drugs, especially TEM-1, has led to a crisis. Now, over 170 different variants of TEM-1 have been isolated in hospitals across the world [6]. Many of these variants contain different phenotypes making them difficult to combat. TEM-1 is commonly used as a standard in experiments to show how antibiotic resistant bacteria have evolved over time. This is partly due to the fact of TEM-1 being first documented in the 1960s [7].
Structural highlights
This TEM1 Class Antibiotic Resistant protein is similar in structure to other Beta Lactamases in its secondary structure. The protein consists of 263 amino acid residues, which are arranged into a particular structure to provide the function it was designed for. The structure itself consists of two main domains, forming a cleft in between them as the active site for the binding of a sulfate anion ligand. The first domain consists of five beta sheets, with three alpha helices overlaying the side facing the solvent, and another alpha helix flanking the sheets on the adjacent side. The other domain is characterized by its high concentration of alpha helices throughout the secondary structure of this domain. The protein in solution is solvated by 135 water molecules per asymmetric unit. In terms of overall structure, 42.2% consists of alpha helices, 17.5% participate in beta sheets, and 37.2% are turns and coils [8]. Lysine 234, which is present on the wall of the active site as a constituent of one of the beta sheets, has been elucidated as an active binding agent, involved in both the recognition of the substrate and stabilization of the intermediate [8]. Furthermore, the amine group of the Lysine residue is involved in electrostatic binding of the C3 carboxylic group of penicillins. Tyrosine 105 has also been elucidated as a binding mechanism for substrates, as its substitution in the active site has proven to cause decreased catalytic activity [9].
References
1. Davies, J.; Davies, G. (2010) Origins and Evolution of Antibiotic Resistance. Microbiol Mol Biol Rev., 74(3): 417–433.
2. National Institute of Health. (2014) Stop the Spread of Superbugs Help Fight Drug-Resistant Bacteria. https://newsinhealth.nih.gov/issue/feb2014/feature1. (Last accessed: April 11, 2016).
3. Dablon et al. (1996) The catalytic mechanism of f3-lactamases: NMR titration of an active-site lysine residue of the TEM-1 enzyme. Proc. Natl. Acad. Sci. USA. 74, 1747-1752.
4. Fonze, E.; Charlier, P.; To'th, Y.; Vermeire, M.; Raquet, X.; Dubus, A.; Frere, J. M. (1995) TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr. D Biol. Crystallogr. 51, 682-694.
5. Salverda, M. L.; Visser, J. A. G. D.; Barlow, M. (2010) Natural Evolution of TEM-1 β-Lactamase: Experimental Reconstruction and Clinical Relevance. FEMS Microbiology Reviews FEMS Microbiol Rev. 34, 1015–1036.
6. Neuwirth, C.; Madec, S.; Siebor, E.; Pechinot, A.; Duez, J.-M.; Pruneaux, M.; Fouchereau-Peron, M.; Kazmierczak, A.; Labia, R. (2010) TEM-89 Beta -Lactamase Produced by a Proteus Mirabilis Clinical Isolate: New Complex Mutant (CMT 3) with Mutations in Both TEM-59 (IRT-17) and TEM-3. Antimicrobial Agents and Chemotherapy 45, 3591–3594.
7. Elander, R. P. (2003) Industrial Production of β-Lactam Antibiotics. Appl Microbiol Biotechnol Applied Microbiology and Biotechnology. 61, 385–392.
8. Lenfant, F.; Labia, R.; Masson, J. (1991) Replacement of lysine 234 affects transition state stabilization in the active site of ß-lactamase TEM1. J. Biol. Chem. 266, 17187-17194.
9. Doucet, N.; Savard, P. -.; Pelletier, J. N.; Gagné, S. M. (2007) NMR investigation of Tyr105 mutants in TEM-1 ß-lactamase: Dynamics are correlated with function. J. Biol. Chem. 282, 21448-21459.



