User:R. Jeremy Johnson/Lysophosphatidic acid receptor 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
| - | Lysophosphatidic Acid Receptor 1 (commonly referred to as LPA<sub>1</sub>) is a [[G protein-coupled receptor]] and one of 6 different LPA receptors (LPA<sub>1</sub>-LPA<sub>6</sub>). These receptors bind the phospholipid derivative [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a signaling molecule that acts as a potent [https://en.wikipedia.org/wiki/Mitogen mitogen] upon binding to one of its six receptors.<ref name="regpeps">PMID: 26091040</ref> LPA<sub>1</sub> is part of the larger [http://jb.oxfordjournals.org/content/131/6/767 EDG receptor family], which includes the more widely studied sphingosine 1-phopshate receptors.<ref name="regpeps">PMID: 26091040</ref> This receptor is responsible for initiating several different signaling cascades with different molecules and G-proteins.<ref name = 'Yung'>Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.' </ref> These cascades ultimately result in growth, survival, and movement of cells, as well as neural cell development.<ref name = 'Chun'>Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.' </ref> | + | Lysophosphatidic Acid Receptor 1 (commonly referred to as LPA<sub>1</sub>) is a [[G protein-coupled receptor]] and one of 6 different LPA receptors (LPA<sub>1</sub>-LPA<sub>6</sub>). These receptors bind the phospholipid derivative [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a signaling molecule that acts as a potent [https://en.wikipedia.org/wiki/Mitogen mitogen] upon binding to one of its six receptors.<ref name="regpeps">PMID: 26091040</ref> LPA<sub>1</sub> is part of the larger [http://jb.oxfordjournals.org/content/131/6/767 EDG receptor family], which includes the more widely studied sphingosine 1-phopshate receptors.<ref name="regpeps">PMID: 26091040</ref> This receptor is responsible for initiating several different signaling cascades with different molecules and G-proteins.<ref name = 'Yung'>Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.' </ref> These cascades ultimately result in growth, survival, and movement of cells, as well as neural cell development.<ref name = 'Chun'>Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.' </ref> |
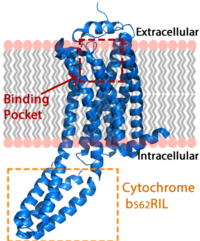
| + | [[Image:LPA_in_membrane4.fw.png|200px|center|thumb|'''Figure 1:''' LPA receptor (blue) bound to the cell membrane. The binding pocket is highlighted in red. The added bRIL protein is highlighted in orange.]] | ||
== Lysophosphatidic Acid == | == Lysophosphatidic Acid == | ||
| Line 9: | Line 10: | ||
Extracellularly, LPA is produced from lysophosphatidylcholine by the enzyme autotaxin.<ref name= "Chrencik"/> Autotaxin was originally linked with metastasis, and this link was later discovered to be mediated through the production of LPA, which signals cell proliferation.<ref name= "Boutin"> DOI: 10.1007/s00018-009-0056-9 </ref> All of LPA’s activities are receptor mediated; the signalling lipid interacts with at least six G-protein coupled receptors LPA<sub>1</sub>-LPA<sub>6</sub>. | Extracellularly, LPA is produced from lysophosphatidylcholine by the enzyme autotaxin.<ref name= "Chrencik"/> Autotaxin was originally linked with metastasis, and this link was later discovered to be mediated through the production of LPA, which signals cell proliferation.<ref name= "Boutin"> DOI: 10.1007/s00018-009-0056-9 </ref> All of LPA’s activities are receptor mediated; the signalling lipid interacts with at least six G-protein coupled receptors LPA<sub>1</sub>-LPA<sub>6</sub>. | ||
| + | |||
== Structure == | == Structure == | ||
| Line 30: | Line 32: | ||
=== Sphingosine 1-Phosphate Receptor === | === Sphingosine 1-Phosphate Receptor === | ||
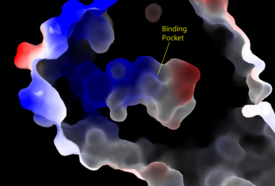
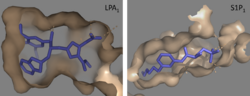
| - | Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>. <ref name= "Chrencik"/> A major difference was observed in ligand access between these two receptors. The binding path in LPA<sub>1</sub> is located in the extracellular milieu, while in S1P<sub>1</sub> the ligand accesses the binding pocket through the membrane (Figure 3). The overall shape of each binding pocket is also different, as the S1P<sub>1</sub> binding pocket has more of an oval shape, whereas [[Image:LPA S1P.png| | + | Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>. <ref name= "Chrencik"/> A major difference was observed in ligand access between these two receptors. The binding path in LPA<sub>1</sub> is located in the extracellular milieu, while in S1P<sub>1</sub> the ligand accesses the binding pocket through the membrane (Figure 3). The overall shape of each binding pocket is also different, as the S1P<sub>1</sub> binding pocket has more of an oval shape, whereas [[Image:LPA S1P.png|250px|right|thumb|'''Figure 5:''' Comparison of the binding pockets of LPA<sub>1</sub> and S1P<sub>1</sub> receptors. The electron density (tan) of the binding pocket is shown around the ligand (purple). The limited binding sites of the receptors are shown in tan.]] the LPA<sub>1</sub> binding pocket has a more spherical shape (Figure 5). The more spherical binding pocket for LPA<sub>1</sub> also gives it the ability to recognize a larger group of chemical species. In particular, LPA<sub>1</sub> has the ability to bind with ligands that have acyl chains of varying lengths <ref name= "Chrencik"/>. Since LPA<sub>1</sub> binds with a variety of acyl chains, it can be used in multiple pathways. |
Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps">PMID: 26091040</ref> This narrowing of the gap between TMI and TMVII blocks membrane ligand access in LPA<sub>1</sub>, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results from ECL0 lack of secondary structure in LPA<sub>1</sub> further promotes favorable LPA access to the binding pocket from the extracellular space. <ref name="regpeps">PMID: 26091040</ref> | Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps">PMID: 26091040</ref> This narrowing of the gap between TMI and TMVII blocks membrane ligand access in LPA<sub>1</sub>, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results from ECL0 lack of secondary structure in LPA<sub>1</sub> further promotes favorable LPA access to the binding pocket from the extracellular space. <ref name="regpeps">PMID: 26091040</ref> | ||
| Line 52: | Line 54: | ||
=== Fibrosis === | === Fibrosis === | ||
Idiopathic pulmonary fibrosis (IPF) has high rates of mortality <ref name= "Tager"> PMID:18066075 </ref>. Understanding how LPA can effect fibrosis, is an important factor to finding medication and a cure for this disease. The pathway of LPA-LPA<sub>1</sub> is important in mediating fibroblast migration and [https://en.wikipedia.org/wiki/Wound_healing Wound Healing]. Once fibrosis has been contracted LPA levels increase in the bronchoalveolar lavage (BAL) fluid. The study showed that mice lacking LPA<sub>1</sub> had protection from mortality and were able to survive fibrosis. LPA<sub>1</sub> plays an active role between lung injury and contracting pulmonary fibrosis. The absence of LPA results in a vascular leak after an initial injury, leading to fibrosis. LPA<sub>1</sub> is a link between lung injury and [http://www.nature.com/nm/journal/v14/n1/fig_tab/nm1685_F4.html pulmonary fibrosis] <ref name= "Tager"/>. | Idiopathic pulmonary fibrosis (IPF) has high rates of mortality <ref name= "Tager"> PMID:18066075 </ref>. Understanding how LPA can effect fibrosis, is an important factor to finding medication and a cure for this disease. The pathway of LPA-LPA<sub>1</sub> is important in mediating fibroblast migration and [https://en.wikipedia.org/wiki/Wound_healing Wound Healing]. Once fibrosis has been contracted LPA levels increase in the bronchoalveolar lavage (BAL) fluid. The study showed that mice lacking LPA<sub>1</sub> had protection from mortality and were able to survive fibrosis. LPA<sub>1</sub> plays an active role between lung injury and contracting pulmonary fibrosis. The absence of LPA results in a vascular leak after an initial injury, leading to fibrosis. LPA<sub>1</sub> is a link between lung injury and [http://www.nature.com/nm/journal/v14/n1/fig_tab/nm1685_F4.html pulmonary fibrosis] <ref name= "Tager"/>. | ||
| - | |||
| - | StructureSection load='4z34' size='340' side='right' caption=' LPA Receptor 1 ' scene='72/721545/Overall/2' | ||
== References == | == References == | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||