User:R. Jeremy Johnson/Lysophosphatidic acid receptor 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
| - | Lysophosphatidic Acid Receptor 1 (commonly referred to as LPA<sub>1</sub>) is a [[G protein-coupled receptor]] and one of 6 different LPA receptors (LPA<sub>1</sub>-LPA<sub>6</sub>). These receptors bind the phospholipid derivative [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a signaling molecule that acts as a potent [https://en.wikipedia.org/wiki/Mitogen mitogen] upon binding to one of its six receptors.<ref name="regpeps">PMID: 26091040</ref> LPA<sub>1</sub> is part of the larger [http://jb.oxfordjournals.org/content/131/6/767 EDG receptor family], which includes the more widely studied sphingosine 1-phopshate receptors.<ref name="regpeps" | + | Lysophosphatidic Acid Receptor 1 (commonly referred to as LPA<sub>1</sub>) is a [[G protein-coupled receptor]] and one of 6 different LPA receptors (LPA<sub>1</sub>-LPA<sub>6</sub>). These receptors bind the phospholipid derivative [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a signaling molecule that acts as a potent [https://en.wikipedia.org/wiki/Mitogen mitogen] upon binding to one of its six receptors.<ref name="regpeps">PMID: 26091040</ref> LPA<sub>1</sub> is part of the larger [http://jb.oxfordjournals.org/content/131/6/767 EDG receptor family], which includes the more widely studied sphingosine 1-phopshate receptors.<ref name="regpeps"/> This receptor is responsible for initiating several different signaling cascades with different molecules and G-proteins.<ref name = 'Yung'>Yung, Y. C., N. C. Stoddard, and J. Chun. "LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology." The Journal of Lipid Research 55.7 (2014): 1192-214. Web. 17 Feb. 2016.' </ref> These cascades ultimately result in growth, survival, and movement of cells, as well as neural cell development.<ref name = 'Chun'>Chun, J., Hla, T., Spiegel, S., and Moolenaar, W.H. “Lysophospholipid Receptors: Signaling and Biochemistry.” John Wiley & Sons, Inc. (2013) pp.i-xviii. 5 Feb. 2016.' </ref> |
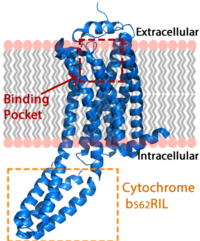
[[Image:LPA_in_membrane4.fw.png|200px|center|thumb|'''Figure 1:''' LPA receptor (blue) bound to the cell membrane. The binding pocket is highlighted in red. The added bRIL protein is highlighted in orange.]] | [[Image:LPA_in_membrane4.fw.png|200px|center|thumb|'''Figure 1:''' LPA receptor (blue) bound to the cell membrane. The binding pocket is highlighted in red. The added bRIL protein is highlighted in orange.]] | ||
| Line 13: | Line 13: | ||
== Structure == | == Structure == | ||
| - | The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> Within these helices is an interior binding pocket that stabilizes the binding of LPA<sub>1</sub>'s natural ligand, LPA (Figure 1). The opening to this binding pocket is larger than other LPA receptors, enabling this receptor to bind ligands other than its natural ligand, such as [https://en.wikipedia.org/wiki/2-Arachidonoylglycerol 2-AG].<ref name="regpeps" | + | The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> Within these helices is an interior binding pocket that stabilizes the binding of LPA<sub>1</sub>'s natural ligand, LPA (Figure 1). The opening to this binding pocket is larger than other LPA receptors, enabling this receptor to bind ligands other than its natural ligand, such as [https://en.wikipedia.org/wiki/2-Arachidonoylglycerol 2-AG].<ref name="regpeps"/> |
LPA<sub>1</sub> lies in the membrane as shown in Figure 1, and as shown by the <scene name='72/721545/Membrane/6'>fatty acid</scene> bound in the crystallization of LPA<sub>1</sub> in orange. Most <scene name='72/721545/Polarity/4'>polar amino acids</scene> (red) reside on the intracellular and extracellular areas of the receptor, while most residues positioned on the trans membrane helices inside the membrane are hydrophobic (blue). A cytochrome b (b<sub>562</sub>RIL) protein was inserted into the third intracellular loop to facilitate crystallization (Figure 2).<ref name= "Chrencik"/> The intracellular region of this membrane protein is coupled to a [https://www.ebi.ac.uk/interpro/potm/2004_10/Page2.htm heterotrimeric G protein]. | LPA<sub>1</sub> lies in the membrane as shown in Figure 1, and as shown by the <scene name='72/721545/Membrane/6'>fatty acid</scene> bound in the crystallization of LPA<sub>1</sub> in orange. Most <scene name='72/721545/Polarity/4'>polar amino acids</scene> (red) reside on the intracellular and extracellular areas of the receptor, while most residues positioned on the trans membrane helices inside the membrane are hydrophobic (blue). A cytochrome b (b<sub>562</sub>RIL) protein was inserted into the third intracellular loop to facilitate crystallization (Figure 2).<ref name= "Chrencik"/> The intracellular region of this membrane protein is coupled to a [https://www.ebi.ac.uk/interpro/potm/2004_10/Page2.htm heterotrimeric G protein]. | ||
| Line 23: | Line 23: | ||
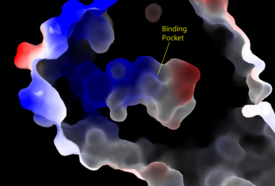
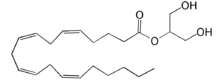
[[Image:Amphbindingfinal.png|275 px|right|thumb|'''Figure 3''': Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] The biological ligand of the LPA<sub>1</sub> receptor receptor is [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a phospholipid that contains a long, nonpolar tail, a phosphate head, a chiral hydroxyl group, and an ester group. This receptor provides specificity for its ligand by the amphipathic binding pocket; the positive region on the left hand side of the pocket stabilizes the LPA's phosphate group, the nonpolar region at the bottom of the binding pocket stabilizes the hydrophobic tail of LPA, and the polar region at the top of the pocket stabilize binding of the ester and hydroxyl group (Figure 3). The <scene name='72/721545/Ligand/4'>binding pocket</scene> for LPA consists of both polar and nonpolar residues. <scene name='72/721545/All_polar_interactions/7'>Polar</scene> residues are located on the N terminus and within the binding pocket. A <scene name='72/721545/Hydrophobic_pocket/4'>hydrophobic pocket</scene> also interacts with the long acyl chain of LPA. The shape and polarity of the binding pocket makes it specific for molecules with a polar head and long hydrophobic tail shaped like LPA. | [[Image:Amphbindingfinal.png|275 px|right|thumb|'''Figure 3''': Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] The biological ligand of the LPA<sub>1</sub> receptor receptor is [https://en.wikipedia.org/wiki/Lysophosphatidic_acid lysophosphatidic acid (LPA)], a phospholipid that contains a long, nonpolar tail, a phosphate head, a chiral hydroxyl group, and an ester group. This receptor provides specificity for its ligand by the amphipathic binding pocket; the positive region on the left hand side of the pocket stabilizes the LPA's phosphate group, the nonpolar region at the bottom of the binding pocket stabilizes the hydrophobic tail of LPA, and the polar region at the top of the pocket stabilize binding of the ester and hydroxyl group (Figure 3). The <scene name='72/721545/Ligand/4'>binding pocket</scene> for LPA consists of both polar and nonpolar residues. <scene name='72/721545/All_polar_interactions/7'>Polar</scene> residues are located on the N terminus and within the binding pocket. A <scene name='72/721545/Hydrophobic_pocket/4'>hydrophobic pocket</scene> also interacts with the long acyl chain of LPA. The shape and polarity of the binding pocket makes it specific for molecules with a polar head and long hydrophobic tail shaped like LPA. | ||
| - | [http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=8589 ONO-9780307 (ON7)] is an antagonist for LPA due to its large nonpolar region, chiral hydroxyl group, ester, and carboxylic acid which all resemble portions of the LPA molecule (Figure 3). Four separate interactions with this antagonist of LPA<sub>1</sub> help demonstrate the key interactions that stabilize the binding of the LPA phospholipid to this receptor. In the nonpolar region of the binding pocket, <scene name='72/721543/Nonpolar/2'>three non polar residues</scene> of LPA<sub>1</sub> stabilize the large nonpolar group of ON7. At the polar region, the ligand binding is stabilized by <scene name='72/721543/Arg124gln125/4'>Arg124 and Glu125</scene> forming ionic and polar interactions with the carboxylic acid and the hydroxyl group of ON7.<ref name="regpeps" | + | [http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=8589 ONO-9780307 (ON7)] is an antagonist for LPA due to its large nonpolar region, chiral hydroxyl group, ester, and carboxylic acid which all resemble portions of the LPA molecule (Figure 3). Four separate interactions with this antagonist of LPA<sub>1</sub> help demonstrate the key interactions that stabilize the binding of the LPA phospholipid to this receptor. In the nonpolar region of the binding pocket, <scene name='72/721543/Nonpolar/2'>three non polar residues</scene> of LPA<sub>1</sub> stabilize the large nonpolar group of ON7. At the polar region, the ligand binding is stabilized by <scene name='72/721543/Arg124gln125/4'>Arg124 and Glu125</scene> forming ionic and polar interactions with the carboxylic acid and the hydroxyl group of ON7.<ref name="regpeps"/> In addition, interplay between <scene name='72/721543/Lys39_and_glu293/8'>Glu293 and Lys39</scene> causes another stabilizing component with the ON7 antagonist. Glu293 forms polar interactions with Lys39, positioning it in close proximity to to the carboxylic acid of ON7, which then interactions with Lys39 via ionic bonding.<ref name="regpeps"/> While Lys39 is highly conserved among all six LPA receptors, a neighboring histidine residue is specific to the LPA<sub>1</sub> receptor. <scene name='72/721543/His40/4'>His40</scene> forms both ionic and polar interactions with the carboxylic acid of ON7. Protonation of this residue greatly affects the binding affinity of LPA, leading to an increase in the pathways associated with cell proliferation and migration. Because cancerous tumors create acidic environments where His40 is protonated, this residue is an important link to tumor growth and cancer cell movement.<ref name="regpeps"/> |
[[Image:Compare.png|175 px|left|thumb|'''Figure 4''': Structures of LPA and its antagonist, ON7.]] | [[Image:Compare.png|175 px|left|thumb|'''Figure 4''': Structures of LPA and its antagonist, ON7.]] | ||
| Line 34: | Line 34: | ||
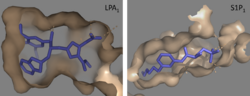
Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>. <ref name= "Chrencik"/> A major difference was observed in ligand access between these two receptors. The binding path in LPA<sub>1</sub> is located in the extracellular milieu, while in S1P<sub>1</sub> the ligand accesses the binding pocket through the membrane (Figure 3). The overall shape of each binding pocket is also different, as the S1P<sub>1</sub> binding pocket has more of an oval shape, whereas [[Image:LPA S1P.png|250px|right|thumb|'''Figure 5:''' Comparison of the binding pockets of LPA<sub>1</sub> and S1P<sub>1</sub> receptors. The electron density (tan) of the binding pocket is shown around the ligand (purple). The limited binding sites of the receptors are shown in tan.]] the LPA<sub>1</sub> binding pocket has a more spherical shape (Figure 5). The more spherical binding pocket for LPA<sub>1</sub> also gives it the ability to recognize a larger group of chemical species. In particular, LPA<sub>1</sub> has the ability to bind with ligands that have acyl chains of varying lengths <ref name= "Chrencik"/>. Since LPA<sub>1</sub> binds with a variety of acyl chains, it can be used in multiple pathways. | Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>. <ref name= "Chrencik"/> A major difference was observed in ligand access between these two receptors. The binding path in LPA<sub>1</sub> is located in the extracellular milieu, while in S1P<sub>1</sub> the ligand accesses the binding pocket through the membrane (Figure 3). The overall shape of each binding pocket is also different, as the S1P<sub>1</sub> binding pocket has more of an oval shape, whereas [[Image:LPA S1P.png|250px|right|thumb|'''Figure 5:''' Comparison of the binding pockets of LPA<sub>1</sub> and S1P<sub>1</sub> receptors. The electron density (tan) of the binding pocket is shown around the ligand (purple). The limited binding sites of the receptors are shown in tan.]] the LPA<sub>1</sub> binding pocket has a more spherical shape (Figure 5). The more spherical binding pocket for LPA<sub>1</sub> also gives it the ability to recognize a larger group of chemical species. In particular, LPA<sub>1</sub> has the ability to bind with ligands that have acyl chains of varying lengths <ref name= "Chrencik"/>. Since LPA<sub>1</sub> binds with a variety of acyl chains, it can be used in multiple pathways. | ||
| - | Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps" | + | Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps"/> This narrowing of the gap between TMI and TMVII blocks membrane ligand access in LPA<sub>1</sub>, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results from ECL0 lack of secondary structure in LPA<sub>1</sub> further promotes favorable LPA access to the binding pocket from the extracellular space. <ref name="regpeps"/> |
=== Endocannabinoid Receptor 1 === | === Endocannabinoid Receptor 1 === | ||
| - | LPA<sub>1</sub> is also closely related to the first of the six [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and LPA<sub>1</sub> the ability to bind to analogs of CB<sub>1</sub> ligands. <ref name="regpeps" | + | LPA<sub>1</sub> is also closely related to the first of the six [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and LPA<sub>1</sub> the ability to bind to analogs of CB<sub>1</sub> ligands. <ref name="regpeps"/> This crossing over of ligand binding opens the possibility of metabolic crosstalk between the two signaling systems. <ref name="regpeps"/> Complementary access to the LPA<sub>1</sub> binding pocket can be achieved by phosphorylated CB<sub>1</sub> ligand analogs, while complementary access to the CB<sub>1</sub> binding site requires dephosphorylation of LPA<sub>1</sub> ligand analogs. In both cases, a ligand could serve as a primary [https://en.wikipedia.org/wiki/Selective_receptor_modulator receptor modulator] and a simultaneous [https://en.wikipedia.org/wiki/Prodrug prodrug] for a different receptor. <ref name="regpeps"/> |
A major cannabinoid signaling molecule, 2-arachidonyl glycerol (2-AG, Figure 5), can be phosphorylated into 2-arachidonyl phosphatidic acid (2-ALPA). 2-ALPA has a similar structure to LPA, and is able to bind in the LPA<sub>1</sub> receptor binding pocket. 2-ALPA binding to LPA<sub>1</sub> causes the same downstream signaling that the LPA molecule does, effectively connecting these two systems. Promiscuous ligand binding between these two pathways has potential functional and therapeutic implications.<ref name= "Chrencik"/>[[Image:2-AG.png|220px|right|thumb|'''Figure 5:''' 2-arachidonylglycerol (2-AG)]] | A major cannabinoid signaling molecule, 2-arachidonyl glycerol (2-AG, Figure 5), can be phosphorylated into 2-arachidonyl phosphatidic acid (2-ALPA). 2-ALPA has a similar structure to LPA, and is able to bind in the LPA<sub>1</sub> receptor binding pocket. 2-ALPA binding to LPA<sub>1</sub> causes the same downstream signaling that the LPA molecule does, effectively connecting these two systems. Promiscuous ligand binding between these two pathways has potential functional and therapeutic implications.<ref name= "Chrencik"/>[[Image:2-AG.png|220px|right|thumb|'''Figure 5:''' 2-arachidonylglycerol (2-AG)]] | ||
| - | <scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains, including the ligand LPA. The polarity of these residues provide favorable interactions between the ligand and the binding pocket.<ref name="regpeps" | + | <scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains, including the ligand LPA. The polarity of these residues provide favorable interactions between the ligand and the binding pocket.<ref name="regpeps"/> Additionally, Asp129 and Trp210 may serve as a trigger for agonist induced conformational changes. These residues are also interesting in regard to GPCR phylogenic evolution. <ref name="regpeps"/> The polar amino acid <scene name='72/721545/210/1'>Trp210</scene> in the binding pocket of LPA<sub>1</sub> is unique to the lysophospholipid and cannabinoid receptors, suggesting that they are related. A model for lipid agonist binding generated through molecular modeling was used to dock two of the cannabinoid receptor CB<sub>1</sub>'s most abundant endogenous ligands into the LPA<sub>1</sub> binding pocket. <ref name="regpeps"/> Rotameric shifts of Trp210 and Trp271 lead to the expansion of the binding pocket and the exposure of the π clouds of their indole rings. These shifts and expansion provided favorable interactions with the double bonds of the phosphorylated cannabinoid ligands. This favorable binding provided evidence that the hydrophobic binding pockets of LPA<sub>1</sub> and CB<sub>1</sub> are able to favorably bind the same poly-unsaturated acyl chains with metabolically interconvertible head groups.<ref name="regpeps"/> |
== Disease Relevance == | == Disease Relevance == | ||
| Line 47: | Line 47: | ||
=== Cancer === | === Cancer === | ||
| - | Many of the functions of LPA<sub>1</sub>, i.e. cell proliferation, survival, and morphology, are implicated in cancers. LPA acts as a tumor mitogen and an inducer of tumor-derived cytokine to support the metastasis (spreading) of breast and ovarian cancer to bones. Inhibition of LPA<sub>1</sub> can significantly reduce this progression, and therefore may be a promising treatment for patients with bone metastasis. <ref name= "Boucharaba"> DOI: 10.1073/pnas.0600979103 </ref> LPA does not have an effect on primary tumor size. <ref name= "Jean > DOI: 10.1093/jnci/djs319 </ref> Part of this tumorigenesis can be explained by the action of <scene name='72/721543/His40/4'>His40</scene> on LPA<sub>1</sub> where, when protonated, increases LPA binding affinity by up to 1kcal/mol.<ref name="regpeps" | + | Many of the functions of LPA<sub>1</sub>, i.e. cell proliferation, survival, and morphology, are implicated in cancers. LPA acts as a tumor mitogen and an inducer of tumor-derived cytokine to support the metastasis (spreading) of breast and ovarian cancer to bones. Inhibition of LPA<sub>1</sub> can significantly reduce this progression, and therefore may be a promising treatment for patients with bone metastasis. <ref name= "Boucharaba"> DOI: 10.1073/pnas.0600979103 </ref> LPA does not have an effect on primary tumor size. <ref name= "Jean > DOI: 10.1093/jnci/djs319 </ref> Part of this tumorigenesis can be explained by the action of <scene name='72/721543/His40/4'>His40</scene> on LPA<sub>1</sub> where, when protonated, increases LPA binding affinity by up to 1kcal/mol.<ref name="regpeps"/> Consequently, in the acidic environment produced by [https://en.wikipedia.org/wiki/Tumor_hypoxia hypoxic tumors] creating lactic acid, LPA<sub>1</sub> activity is increased, allowing these tumors to continue to proliferate, migrate, and survive.<ref name="number7">PMID: 24367336</ref> |
===Pain=== | ===Pain=== | ||