User:R. Jeremy Johnson/Lysophosphatidic acid receptor 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Lysophosphatidic Acid == | == Lysophosphatidic Acid == | ||
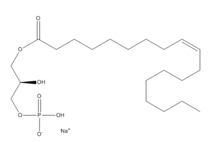
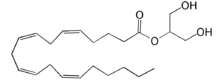
| - | [[Image:LPA.png|220px|left|thumb|'''Figure 2:''' Chemical Structure of LPA (monoacyl-sn-glycero-3-phosphate)]] Lysophosphatidic Acid (LPA) consists of an unsaturated fatty acid chain, a glycerol backbone, and a free phosphate group (Figure 2). Lysophosphatidic acid is found in nearly all cells, tissues, and fluids of the body.<ref name= " | + | [[Image:LPA.png|220px|left|thumb|'''Figure 2:''' Chemical Structure of LPA (monoacyl-sn-glycero-3-phosphate)]] Lysophosphatidic Acid (LPA) consists of an unsaturated fatty acid chain, a glycerol backbone, and a free phosphate group (Figure 2). Lysophosphatidic acid is found in nearly all cells, tissues, and fluids of the body.<ref name="regpeps"/> LPA is present intracellularly as a precursor of phospholipid biosynthesis, and extracellularly as a signalling phospholipid. |
| - | Extracellularly, LPA is produced from lysophosphatidylcholine by the enzyme autotaxin.<ref name= " | + | Extracellularly, LPA is produced from lysophosphatidylcholine by the enzyme autotaxin.<ref name="regpeps"/> Autotaxin was originally linked with metastasis, and this link was later discovered to be mediated through the production of LPA, which signals cell proliferation.<ref name= "Boutin"> DOI: 10.1007/s00018-009-0056-9 </ref> All of LPA’s activities are receptor mediated; the signalling lipid interacts with at least six G-protein coupled receptors LPA<sub>1</sub>-LPA<sub>6</sub>. |
| Line 15: | Line 15: | ||
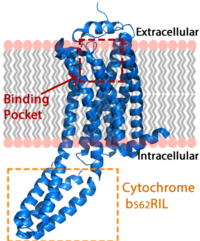
The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> Within these helices is an interior binding pocket that stabilizes the binding of LPA<sub>1</sub>'s natural ligand, LPA (Figure 1). The opening to this binding pocket is larger than other LPA receptors, enabling this receptor to bind ligands other than its natural ligand, such as [https://en.wikipedia.org/wiki/2-Arachidonoylglycerol 2-AG].<ref name="regpeps"/> | The LPA<sub>1</sub> receptor protein is composed of 364 amino acids with a molecular weight of approximately 41 kDa. Common to all G-protein coupled receptors, LPA<sub>1</sub> contains seven [http://kinemage.biochem.duke.edu/teaching/anatax/html/anatax.2a.html alpha helices] which make up the seven transmembrane spanning domains with three intracellular loops and three extracellular loops.<ref name = 'Hernández-Méndez'>Hernández-Méndez, Aurelio, Rocío Alcántara-Hernández, and J. Adolfo García-Sáinz. "Lysophosphatidic Acid LPA1-3 Receptors: Signaling, Regulation and in Silico Analysis of Their Putative Phosphorylation Sites." Receptors & Clinical Investigation Receptor Clin Invest 1.3 (2014). Web. 15 Feb. 2016.' </ref> Within these helices is an interior binding pocket that stabilizes the binding of LPA<sub>1</sub>'s natural ligand, LPA (Figure 1). The opening to this binding pocket is larger than other LPA receptors, enabling this receptor to bind ligands other than its natural ligand, such as [https://en.wikipedia.org/wiki/2-Arachidonoylglycerol 2-AG].<ref name="regpeps"/> | ||
| - | LPA<sub>1</sub> lies in the membrane as shown in Figure 1, and as shown by the <scene name='72/721545/Membrane/6'>fatty acid</scene> bound in the crystallization of LPA<sub>1</sub> in orange. Most <scene name='72/721545/Polarity/4'>polar amino acids</scene> (red) reside on the intracellular and extracellular areas of the receptor, while most residues positioned on the trans membrane helices inside the membrane are hydrophobic (blue). A cytochrome b (b<sub>562</sub>RIL) protein was inserted into the third intracellular loop to facilitate crystallization (Figure 2).<ref name= " | + | LPA<sub>1</sub> lies in the membrane as shown in Figure 1, and as shown by the <scene name='72/721545/Membrane/6'>fatty acid</scene> bound in the crystallization of LPA<sub>1</sub> in orange. Most <scene name='72/721545/Polarity/4'>polar amino acids</scene> (red) reside on the intracellular and extracellular areas of the receptor, while most residues positioned on the trans membrane helices inside the membrane are hydrophobic (blue). A cytochrome b (b<sub>562</sub>RIL) protein was inserted into the third intracellular loop to facilitate crystallization (Figure 2).<ref name="regpeps"/> The intracellular region of this membrane protein is coupled to a [https://www.ebi.ac.uk/interpro/potm/2004_10/Page2.htm heterotrimeric G protein]. |
=== Structural Stabilization === | === Structural Stabilization === | ||
| - | Three native <scene name='72/721545/Disulfides/5'>disulfide bonds</scene> in the extracellular region of this receptor provide fold stability.<ref name= " | + | Three native <scene name='72/721545/Disulfides/5'>disulfide bonds</scene> in the extracellular region of this receptor provide fold stability.<ref name="regpeps"/> The first disulfide bond constrains the N terminal helix to extracellular loop(ECL) 2. The second disulfide bond shapes ECL2, and the third binds ECL3 to one of the transmembrane alpha helices. These disulfide bonds provide intramolecular stabilization along the extracellular region of the LPA<sub>1</sub> receptor, where the substrate enters into the binding pocket. The <scene name='72/721545/N-terminus/3'>N-terminus</scene> is a six turn alpha helix. It functions like a cap on the extracellular side of the protein, packing tightly against ECL1 and ECL2.<ref name="regpeps"/> The N-terminus helix also provides <scene name='72/721545/34_39_40/4'>polar amino acids</scene> that interact with the ligand when bound. The extracellular region of this receptor plays a role in substrate specificity. |
=== Key Ligand Interactions === | === Key Ligand Interactions === | ||
| Line 32: | Line 32: | ||
=== Sphingosine 1-Phosphate Receptor === | === Sphingosine 1-Phosphate Receptor === | ||
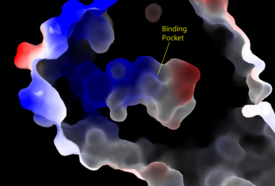
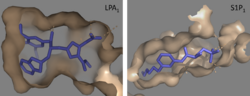
| - | Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>. <ref name= " | + | Lysophosphatidic Acid Receptors (LPA) are part of a larger family known as lysophospholipid receptor family ([http://jb.oxfordjournals.org/content/131/6/767.long EDG family]), including the archetype sphingosine-1-phosphate receptors (S1P<sub>1</sub>). The only structure previously reported in this GPCR family was of S1P<sub>1</sub>, and it provides a comparison for differential structure and function to LPA<sub>1</sub>.<ref name="regpeps"/> A major difference was observed in ligand access between these two receptors. The binding path in LPA<sub>1</sub> is located in the extracellular milieu, while in S1P<sub>1</sub> the ligand accesses the binding pocket through the membrane (Figure 3). The overall shape of each binding pocket is also different, as the S1P<sub>1</sub> binding pocket has more of an oval shape, whereas [[Image:LPA S1P.png|250px|right|thumb|'''Figure 5:''' Comparison of the binding pockets of LPA<sub>1</sub> and S1P<sub>1</sub> receptors. The electron density (tan) of the binding pocket is shown around the ligand (purple). The limited binding sites of the receptors are shown in tan.]] the LPA<sub>1</sub> binding pocket has a more spherical shape (Figure 5). The more spherical binding pocket for LPA<sub>1</sub> also gives it the ability to recognize a larger group of chemical species. In particular, LPA<sub>1</sub> has the ability to bind with ligands that have acyl chains of varying lengths.<ref name="regpeps"/> Since LPA<sub>1</sub> binds with a variety of acyl chains, it can be used in multiple pathways. |
Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps"/> This narrowing of the gap between TMI and TMVII blocks membrane ligand access in LPA<sub>1</sub>, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results from ECL0 lack of secondary structure in LPA<sub>1</sub> further promotes favorable LPA access to the binding pocket from the extracellular space. <ref name="regpeps"/> | Structural evidence for this altered ligand binding pathway includes global changes in the positioning of the extracellular loops (ECL) and transmembrane helices (TM). Specifically, a slight divergence of <scene name='72/721543/Tmvii_and_tmi/1'>TMI</scene>, which is positioned 3 Å closer to TMVII compared to S1P<sub>1</sub>, and a repositioning of <scene name='72/721543/Ecl_regions/1'>ECL3</scene>, resulting in a divergence of 8 Å from S1P<sub>1</sub> result in ligand access via the extracellular space. <ref name="regpeps"/> This narrowing of the gap between TMI and TMVII blocks membrane ligand access in LPA<sub>1</sub>, while the greater distance between ECL3 and the other extracellular loops promotes extracellular access for LPA<sub>1</sub>. Additionally, ECL0 is helical in S1P<sub>1</sub>, but <scene name='72/721543/Ecl02ndstructure/1'>lacks secondary structure</scene> in LPA<sub>1</sub>. This increased flexibility that results from ECL0 lack of secondary structure in LPA<sub>1</sub> further promotes favorable LPA access to the binding pocket from the extracellular space. <ref name="regpeps"/> | ||
| Line 39: | Line 39: | ||
LPA<sub>1</sub> is also closely related to the first of the six [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and LPA<sub>1</sub> the ability to bind to analogs of CB<sub>1</sub> ligands. <ref name="regpeps"/> This crossing over of ligand binding opens the possibility of metabolic crosstalk between the two signaling systems. <ref name="regpeps"/> Complementary access to the LPA<sub>1</sub> binding pocket can be achieved by phosphorylated CB<sub>1</sub> ligand analogs, while complementary access to the CB<sub>1</sub> binding site requires dephosphorylation of LPA<sub>1</sub> ligand analogs. In both cases, a ligand could serve as a primary [https://en.wikipedia.org/wiki/Selective_receptor_modulator receptor modulator] and a simultaneous [https://en.wikipedia.org/wiki/Prodrug prodrug] for a different receptor. <ref name="regpeps"/> | LPA<sub>1</sub> is also closely related to the first of the six [http://www.nature.com/ijo/journal/v30/n1s/full/0803272a.html cannabinoid receptors]. This close relation gives CB<sub>1</sub> ([[Cannabinoid Receptor 1]]) the ability to bind to analogs of LPA and LPA<sub>1</sub> the ability to bind to analogs of CB<sub>1</sub> ligands. <ref name="regpeps"/> This crossing over of ligand binding opens the possibility of metabolic crosstalk between the two signaling systems. <ref name="regpeps"/> Complementary access to the LPA<sub>1</sub> binding pocket can be achieved by phosphorylated CB<sub>1</sub> ligand analogs, while complementary access to the CB<sub>1</sub> binding site requires dephosphorylation of LPA<sub>1</sub> ligand analogs. In both cases, a ligand could serve as a primary [https://en.wikipedia.org/wiki/Selective_receptor_modulator receptor modulator] and a simultaneous [https://en.wikipedia.org/wiki/Prodrug prodrug] for a different receptor. <ref name="regpeps"/> | ||
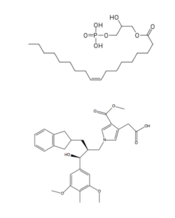
| - | A major cannabinoid signaling molecule, 2-arachidonyl glycerol (2-AG, Figure 5), can be phosphorylated into 2-arachidonyl phosphatidic acid (2-ALPA). 2-ALPA has a similar structure to LPA, and is able to bind in the LPA<sub>1</sub> receptor binding pocket. 2-ALPA binding to LPA<sub>1</sub> causes the same downstream signaling that the LPA molecule does, effectively connecting these two systems. Promiscuous ligand binding between these two pathways has potential functional and therapeutic implications.<ref name= " | + | A major cannabinoid signaling molecule, 2-arachidonyl glycerol (2-AG, Figure 5), can be phosphorylated into 2-arachidonyl phosphatidic acid (2-ALPA). 2-ALPA has a similar structure to LPA, and is able to bind in the LPA<sub>1</sub> receptor binding pocket. 2-ALPA binding to LPA<sub>1</sub> causes the same downstream signaling that the LPA molecule does, effectively connecting these two systems. Promiscuous ligand binding between these two pathways has potential functional and therapeutic implications.<ref name="regpeps"/>[[Image:2-AG.png|220px|right|thumb|'''Figure 5:''' 2-arachidonylglycerol (2-AG)]] |
<scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains, including the ligand LPA. The polarity of these residues provide favorable interactions between the ligand and the binding pocket.<ref name="regpeps"/> Additionally, Asp129 and Trp210 may serve as a trigger for agonist induced conformational changes. These residues are also interesting in regard to GPCR phylogenic evolution. <ref name="regpeps"/> The polar amino acid <scene name='72/721545/210/1'>Trp210</scene> in the binding pocket of LPA<sub>1</sub> is unique to the lysophospholipid and cannabinoid receptors, suggesting that they are related. A model for lipid agonist binding generated through molecular modeling was used to dock two of the cannabinoid receptor CB<sub>1</sub>'s most abundant endogenous ligands into the LPA<sub>1</sub> binding pocket. <ref name="regpeps"/> Rotameric shifts of Trp210 and Trp271 lead to the expansion of the binding pocket and the exposure of the π clouds of their indole rings. These shifts and expansion provided favorable interactions with the double bonds of the phosphorylated cannabinoid ligands. This favorable binding provided evidence that the hydrophobic binding pockets of LPA<sub>1</sub> and CB<sub>1</sub> are able to favorably bind the same poly-unsaturated acyl chains with metabolically interconvertible head groups.<ref name="regpeps"/> | <scene name='72/721543/Asp129_and_trp210/2'>Residues Asp129 and Trp210</scene> located within the hydrophobic binding pocket of LPA<sub>1</sub> may share responsibility for the preference for long unsaturated acyl chains, including the ligand LPA. The polarity of these residues provide favorable interactions between the ligand and the binding pocket.<ref name="regpeps"/> Additionally, Asp129 and Trp210 may serve as a trigger for agonist induced conformational changes. These residues are also interesting in regard to GPCR phylogenic evolution. <ref name="regpeps"/> The polar amino acid <scene name='72/721545/210/1'>Trp210</scene> in the binding pocket of LPA<sub>1</sub> is unique to the lysophospholipid and cannabinoid receptors, suggesting that they are related. A model for lipid agonist binding generated through molecular modeling was used to dock two of the cannabinoid receptor CB<sub>1</sub>'s most abundant endogenous ligands into the LPA<sub>1</sub> binding pocket. <ref name="regpeps"/> Rotameric shifts of Trp210 and Trp271 lead to the expansion of the binding pocket and the exposure of the π clouds of their indole rings. These shifts and expansion provided favorable interactions with the double bonds of the phosphorylated cannabinoid ligands. This favorable binding provided evidence that the hydrophobic binding pockets of LPA<sub>1</sub> and CB<sub>1</sub> are able to favorably bind the same poly-unsaturated acyl chains with metabolically interconvertible head groups.<ref name="regpeps"/> | ||