User:R. Jeremy Johnson/Glucagon Receptor
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
==== Binding Pocket ==== | ==== Binding Pocket ==== | ||
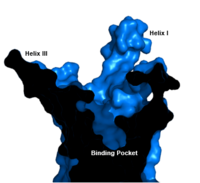
| - | [[Image:Labeled_Binding_Pocket.png|200 px|left|thumb|'''Figure | + | [[Image:Labeled_Binding_Pocket.png|200 px|left|thumb|'''Figure 3. GCGR Binding Pocket.''' A cross-section of the GCGR binding pocket shows its width and depth]]The class B GPCR has the widest and longest <scene name='72/721538/Binding_pocket/1'>binding pocket</scene> of all other classes of GPCRs. The distance between the EC tips of Helicies II and VI as well as between the tips of Helicies III and VII are some of the largest among the GPCRs<ref name="structure_article" />. As a result, the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3820480/bin/nihms495648f2.jpg binding cavity] of GCGR is located deeper inside the receptor, meaning glucagon binds much closer to the cell membrane. |
====Other Unique Structural Features ==== | ====Other Unique Structural Features ==== | ||
| Line 33: | Line 33: | ||
===Glucagon Binding=== | ===Glucagon Binding=== | ||
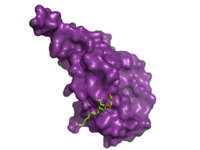
| - | The large, soluble N-terminal extracellular domains (ECD) of GCGR provide initial ligand selectivity with the deep, ligand pocket of the TMD providing secondary recognition.<ref name= "Yang 2015"/> [[Image:ECD_bound_to_glucagon.png|200 px|left|thumb|'''Figure | + | The large, soluble N-terminal extracellular domains (ECD) of GCGR provide initial ligand selectivity with the deep, ligand pocket of the TMD providing secondary recognition.<ref name= "Yang 2015"/> [[Image:ECD_bound_to_glucagon.png|200 px|left|thumb|'''Figure 4. Bound Molecule of Glucagon.''' A molecule of glucagon is shown bound to the GCGR's ECD (shown in magenta)]] The active, or open conformation, is characterized by an intracellular outward movement of <scene name='72/721538/Helix_v_and_vi/1'>helicies V and VI</scene> (breaking hydrogen bonds between <scene name='72/721538/Arg173-ser350_h_bond/1'>Arg173-Ser350</scene> and <scene name='72/721538/Arg173-ser350_h_bond/2'>Glu245-Thr351</scene>)<ref name='therapeutic_article'>DOI 10.1016/j.tips.2013.11.001</ref> and an extracellular rotation of the ECD until it is almost perpendicular to the membrane surface <ref name ='conformation_article'>PMID:26227798 </ref>. While the stalk region of Helix I helps to facilitate the motion of the ECD, intracellular G-protein coupling and extracellular glucagon binding stabilized this active state. In the abscence of glucagon, however, the GCGR adopts a closed conformation in which all three of the extracellular loops of the 7tm (<scene name='72/721538/Ecls/1'>ECL1, ECL2, and ECL3</scene>) can interact with the ECD <ref name ='conformation_article'>PMID:26227798 </ref>. In this closed state, the ECD covers the extracellular surface of the 7tm. To transition between states, the ECD rotates and moves down towards the 7tm domain. This transition mechanism is consistent with the "two-domain" binding mechanism of class B GCPRs in which (1) the C-terminus of the ligand first binds to the ECD allowing (2) the N-terminus of the ligand to interact with the 7tm and activate the protein <ref name='therapeutic_article'>DOI 10.1016/j.tips.2013.11.001</ref>. |
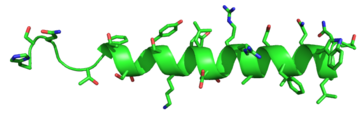
| - | The [https://en.wikipedia.org/wiki/Residue_(chemistry) residues] in the binding pocket that are in direct contact with the glucagon molecule are [https://en.wikipedia.org/wiki/Chemical_polarity polar] (utilizing the attraction of opposite charges or [https://en.wikipedia.org/wiki/Dipole dipoles] for glucagon binding) or are hydrophobic (utilizing the hydrophobic effect). The [https://en.wikipedia.org/wiki/Active_site binding site] location of the hormone peptide ligand has been identified, and the N-terminus of glucagon is known to bind partly with the ECD while the rest of glucagon binds deep into the binding pocket | + | The [https://en.wikipedia.org/wiki/Residue_(chemistry) residues] in the binding pocket that are in direct contact with the glucagon molecule are [https://en.wikipedia.org/wiki/Chemical_polarity polar] (utilizing the attraction of opposite charges or [https://en.wikipedia.org/wiki/Dipole dipoles] for glucagon binding) or are hydrophobic (utilizing the hydrophobic effect). The [https://en.wikipedia.org/wiki/Active_site binding site] location of the hormone peptide ligand has been identified, and the N-terminus of glucagon is known to bind partly with the ECD while the rest of glucagon binds deep into the binding pocket. The [https://en.wikipedia.org/wiki/Amino_acid amino acids] at the N-terminus of the class B 7TM have the ability to form [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonds] and [https://en.wikipedia.org/wiki/Ionic_bonding ionic interactions], which can be seen in the [https://en.wikipedia.org/wiki/Peptide_sequence amino acid sequence] of glucagon (Figure 5). <ref name="Sequence">PMID: 11946536</ref> |
[[Image:Glucagonstructure.png|(|):|360 px|right|thumb|'''Figure 5: Structure of Glucagon:''' The side chains of the residues making up glucagon are depicted. Coloration on the side chains indicate certain [https://en.wikipedia.org/wiki/Atom atoms] that determine the properties the residues hold. The blue indicates a [https://en.wikipedia.org/wiki/Nitrogen nitrogen] atom (hydrophilic properties), the green on the side chains indicates carbon atoms (non-polar hydrophobic properties), and the red coloration indicates an [https://en.wikipedia.org/wiki/Oxygen oxygen] atom (hydrophilic properties). [http://www.rcsb.org/pdb/home/home.do PDB] [http://www.rcsb.org/pdb/explore.do?structureId=1GCN 1GCN] ]] | [[Image:Glucagonstructure.png|(|):|360 px|right|thumb|'''Figure 5: Structure of Glucagon:''' The side chains of the residues making up glucagon are depicted. Coloration on the side chains indicate certain [https://en.wikipedia.org/wiki/Atom atoms] that determine the properties the residues hold. The blue indicates a [https://en.wikipedia.org/wiki/Nitrogen nitrogen] atom (hydrophilic properties), the green on the side chains indicates carbon atoms (non-polar hydrophobic properties), and the red coloration indicates an [https://en.wikipedia.org/wiki/Oxygen oxygen] atom (hydrophilic properties). [http://www.rcsb.org/pdb/home/home.do PDB] [http://www.rcsb.org/pdb/explore.do?structureId=1GCN 1GCN] ]] | ||
Specific amino acid interactions have been identified in GCGR that maximize affinity. This includes the alpha helical structure of the <scene name='72/721535/Stalk/1'>stalk</scene>. The alpha helical structure of the stalk interacts directly with glucagon, as it extends nearly three helical turns above the membrane. When the alpha helix of the stalk is disrupted, the affinity of glucagon for GCGR decreases. A [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] study mutating <scene name='72/721535/Ala135/1'>alanine 135</scene> to a proline. Proline disrupts helices. The Ala135Pro mutant had significant lower affinity for glucagon.<ref name="Tips">PMID: 23863937</ref> Furthermore, there are certain interactions that hold the helices of the 7TM in the conformation that maximizes [http://www.chemicool.com/definition/affinity.html affinity]. <ref name="Ligands">PMID: 21542831</ref> The high affinity conformation of GCGR is the open conformation, when glucagon can bind. Without these specific interactions between the residues, the open conformation is not stabilized and GCGR remains in the closed conformation, where glucagon cannot bind. <ref name="Tips">PMID: 23863937</ref> The [https://en.wikipedia.org/wiki/Disulfide disulfide bond] between <scene name='72/721535/Disulfide_bond_notspin_actual/2'>Cys 294 and Cys 224</scene> serves to hold the helices in the proper orientation for binding and stabilizes the open conformation. Additionally, the [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] between | Specific amino acid interactions have been identified in GCGR that maximize affinity. This includes the alpha helical structure of the <scene name='72/721535/Stalk/1'>stalk</scene>. The alpha helical structure of the stalk interacts directly with glucagon, as it extends nearly three helical turns above the membrane. When the alpha helix of the stalk is disrupted, the affinity of glucagon for GCGR decreases. A [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] study mutating <scene name='72/721535/Ala135/1'>alanine 135</scene> to a proline. Proline disrupts helices. The Ala135Pro mutant had significant lower affinity for glucagon.<ref name="Tips">PMID: 23863937</ref> Furthermore, there are certain interactions that hold the helices of the 7TM in the conformation that maximizes [http://www.chemicool.com/definition/affinity.html affinity]. <ref name="Ligands">PMID: 21542831</ref> The high affinity conformation of GCGR is the open conformation, when glucagon can bind. Without these specific interactions between the residues, the open conformation is not stabilized and GCGR remains in the closed conformation, where glucagon cannot bind. <ref name="Tips">PMID: 23863937</ref> The [https://en.wikipedia.org/wiki/Disulfide disulfide bond] between <scene name='72/721535/Disulfide_bond_notspin_actual/2'>Cys 294 and Cys 224</scene> serves to hold the helices in the proper orientation for binding and stabilizes the open conformation. Additionally, the [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] between | ||
| - | <scene name='72/721535/Salt_bridge_residues/1'>Glu 406, Arg 173, and Arg 346</scene> hold the open conformation together for higher affinity | + | <scene name='72/721535/Salt_bridge_residues/1'>Glu 406, Arg 173, and Arg 346</scene> hold the open conformation together for higher affinity. <ref name="Ligands">PMID: 21542831</ref> |
Mutagenesis and photo cross-linking studies determined essential, conserved residues in glucagon and have been <scene name='72/727091/Glucagon_important_residues/2'>labeled and colored</scene> in red.<ref name= "Siu 2013"/> Glucagon residues His 1, Gln 3, Phe 6, and Tyr 10 are critical to successful binding interaction with the GCGR while others are important for structural rigidity. The n-terminus of glucagon (Fig. 3) leads to a protuberance that fits into the deep, interior cavity of the GCGR 7TMD (Fig. 2) where four residues reside that play strong roles in ligand binding affinity. There is a <scene name='72/721552/Glucagon_binding_zoomed_in/1'>narrow neck</scene> to the entrance of the cavity, providing a firm anchor during peptide docking. (also see Fig. 2) | Mutagenesis and photo cross-linking studies determined essential, conserved residues in glucagon and have been <scene name='72/727091/Glucagon_important_residues/2'>labeled and colored</scene> in red.<ref name= "Siu 2013"/> Glucagon residues His 1, Gln 3, Phe 6, and Tyr 10 are critical to successful binding interaction with the GCGR while others are important for structural rigidity. The n-terminus of glucagon (Fig. 3) leads to a protuberance that fits into the deep, interior cavity of the GCGR 7TMD (Fig. 2) where four residues reside that play strong roles in ligand binding affinity. There is a <scene name='72/721552/Glucagon_binding_zoomed_in/1'>narrow neck</scene> to the entrance of the cavity, providing a firm anchor during peptide docking. (also see Fig. 2) | ||
==Glucagon Signaling Pathway== | ==Glucagon Signaling Pathway== | ||
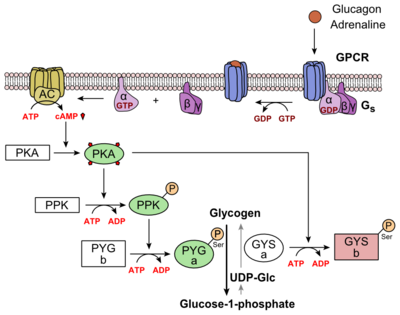
| - | Glucagon binds to the open conformation of GCGR; the GCGR located on the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane]. Glucagon binding to GCGR induces a [https://en.wikipedia.org/wiki/Conformational_change conformational change] in GCGR. This conformation change induces the active state of the protein. The active state of the protein exchanges a [https://en.wikipedia.org/wiki/Guanosine_diphosphate guanosine diphosphate (GDP]) for [https://en.wikipedia.org/wiki/Guanosine_triphosphate guanosine triphosphate (GTP)] that is bound to the [https://en.wikipedia.org/wiki/G_alpha_subunit alpha subunit]. With the GTP in place, the activated alpha subunit dissociates from the [https://en.wikipedia.org/wiki/Heterotrimeric_G_protein heterotrimeric G protein's]beta and gamma subunits. Following dissociation, the alpha subunit can activate [https://en.wikipedia.org/wiki/Adenylyl_cyclase adenylate cyclase]. Activated adenylate cyclase, catalyzes the conversion of [https://en.wikipedia.org/wiki/Adenosine_triphosphate adenosine triphosphate (ATP)] into [https://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate cyclic adenosine monophosphate (cAMP)]. cAMP then serves as a secondary messenger to activate, through allosteric binding, [https://en.wikipedia.org/wiki/Protein_kinase_A cAMP dependent protein kinase A (PKA)]. PKA activates via [https://en.wikipedia.org/wiki/Phosphorylation phosphorylation] the [https://en.wikipedia.org/wiki/Phosphorylase_kinase phosphorylase b kinase]. The phosphorylase b kinase phosphorylates [https://en.wikipedia.org/wiki/Glycogen_phosphorylase glycogen phosphorylase b] to convert to the active form, phosphorylase a. Phosphorylase a finally catalyzes the release of [https://en.wikipedia.org/wiki/Glucose_1-phosphate glucose-1-phosphate] into the bloodstream from glycogen [https://en.wikipedia.org/wiki/Polymer polymers] (Figure | + | Glucagon binds to the open conformation of GCGR; the GCGR located on the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane]. Glucagon binding to GCGR induces a [https://en.wikipedia.org/wiki/Conformational_change conformational change] in GCGR. This conformation change induces the active state of the protein. The active state of the protein exchanges a [https://en.wikipedia.org/wiki/Guanosine_diphosphate guanosine diphosphate (GDP]) for [https://en.wikipedia.org/wiki/Guanosine_triphosphate guanosine triphosphate (GTP)] that is bound to the [https://en.wikipedia.org/wiki/G_alpha_subunit alpha subunit]. With the GTP in place, the activated alpha subunit dissociates from the [https://en.wikipedia.org/wiki/Heterotrimeric_G_protein heterotrimeric G protein's]beta and gamma subunits. Following dissociation, the alpha subunit can activate [https://en.wikipedia.org/wiki/Adenylyl_cyclase adenylate cyclase]. Activated adenylate cyclase, catalyzes the conversion of [https://en.wikipedia.org/wiki/Adenosine_triphosphate adenosine triphosphate (ATP)] into [https://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate cyclic adenosine monophosphate (cAMP)]. cAMP then serves as a secondary messenger to activate, through allosteric binding, [https://en.wikipedia.org/wiki/Protein_kinase_A cAMP dependent protein kinase A (PKA)]. PKA activates via [https://en.wikipedia.org/wiki/Phosphorylation phosphorylation] the [https://en.wikipedia.org/wiki/Phosphorylase_kinase phosphorylase b kinase]. The phosphorylase b kinase phosphorylates [https://en.wikipedia.org/wiki/Glycogen_phosphorylase glycogen phosphorylase b] to convert to the active form, phosphorylase a. Phosphorylase a finally catalyzes the release of [https://en.wikipedia.org/wiki/Glucose_1-phosphate glucose-1-phosphate] into the bloodstream from glycogen [https://en.wikipedia.org/wiki/Polymer polymers] (Figure 6). |
| - | [[Image:Glucagon_Pathway.png|(|):|400 px|center|thumb|'''Figure | + | [[Image:Glucagon_Pathway.png|(|):|400 px|center|thumb|'''Figure 6: [https://en.wikipedia.org/wiki/Glucagon Metabolic Regulation of Glycogen by Glucagon.]'''Depicted is the visualization of the glucagon signaling pathway through the GCGR. The location of the GCGR, the release of the alpha subunit from the beta and gamma subunits, and the enzyme cascade to result in the releasing of glucose are depicted. Abbreviations for the enzymes in the cascade include- PPK: phosphorylase kinase; PYG b: glycogen phosphorylase b; PYG a: glycogen phosphorylase a.]] |
==Clinical relevance == | ==Clinical relevance == | ||
| - | [[Image:Small Molecule Drugs2.jpg|200 px|left|thumb|Figure 4: Three small molecule antagonists reported in 2007.<ref name= "Kazda 2015"/>.]] | ||
Because GCGR can interact with multiple types of G protein subfamilies, discovering small molecule inhibitors could lead to a wide range of focused therapies.<ref name= "Weston 2015"/> Blocking conformations that favor interaction with specific G proteins could allow the knockdown of targeted signal pathways. For example, GCGR is known to interact with inhibitory Gαi proteins that antagonize cAMP production.<ref name= "Weston 2015"/> The finding of an agonist for this pathway could lead to breakthroughs in the treatment of diabetes mellitus. Recently some fundamental work has been done with RAMPs which were shown to alter ligand preference in class B GPCRs.<ref name= "Wootten 2013">DOI:10.1111/j.1476-5381.2012.02202.x</ref> Specifically, RAMP2 association has been shown to alter the pharmacology of all GCGR ligands (glucagon and oxyntomodulin). RAMP2 association altered cAMP production, indicating an effect on signaling bias and g protein coupling.<ref name= "Wootten 2013"/> | Because GCGR can interact with multiple types of G protein subfamilies, discovering small molecule inhibitors could lead to a wide range of focused therapies.<ref name= "Weston 2015"/> Blocking conformations that favor interaction with specific G proteins could allow the knockdown of targeted signal pathways. For example, GCGR is known to interact with inhibitory Gαi proteins that antagonize cAMP production.<ref name= "Weston 2015"/> The finding of an agonist for this pathway could lead to breakthroughs in the treatment of diabetes mellitus. Recently some fundamental work has been done with RAMPs which were shown to alter ligand preference in class B GPCRs.<ref name= "Wootten 2013">DOI:10.1111/j.1476-5381.2012.02202.x</ref> Specifically, RAMP2 association has been shown to alter the pharmacology of all GCGR ligands (glucagon and oxyntomodulin). RAMP2 association altered cAMP production, indicating an effect on signaling bias and g protein coupling.<ref name= "Wootten 2013"/> | ||
| Line 59: | Line 58: | ||
[https://en.wikipedia.org/wiki/G_protein%E2%80%93coupled_receptor G protein-coupled receptors] | [https://en.wikipedia.org/wiki/G_protein%E2%80%93coupled_receptor G protein-coupled receptors] | ||
| - | |||
| - | |||
| - | |||
| - | StructureSection load='4L6R' size='350' side='right' caption='7TM structure of human class B GPCR 4L6R', [[Resolution|resolution]] 1.80Å' scene='72/721552/The_right_one/1' | ||
| - | StructureSection load='4l6r' size='340' side='right' caption='Human Glucagon Class B GPCR ( 7tm PDB: [[4l6r]], ECD PDB: [[4ers]])' scene='72/721538/Glucagon_receptor/2' | ||
</StructureSection> | </StructureSection> | ||
Revision as of 18:51, 9 June 2016
Glucagon G protein-coupled receptor
References
- ↑ 1.0 1.1 1.2 Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006 Feb;2(2):e13. Epub 2006 Feb 17. PMID:16485037 doi:http://dx.doi.org/10.1371/journal.pcbi.0020013
- ↑ 2.0 2.1 2.2 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Bortolato A, Dore AS, Hollenstein K, Tehan BG, Mason JS, Marshall FH. Structure of Class B GPCRs: new horizons for drug discovery. Br J Pharmacol. 2014 Jul;171(13):3132-45. doi: 10.1111/bph.12689. PMID:24628305 doi:http://dx.doi.org/10.1111/bph.12689
- ↑ Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013 Jul 25;499(7459):438-43. doi: 10.1038/nature12357. Epub 2013 Jul 17. PMID:23863939 doi:http://dx.doi.org/10.1038/nature12357
- ↑ Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ 8.0 8.1 8.2 8.3 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859
- ↑ 10.0 10.1 10.2 10.3 Miller LJ, Dong M, Harikumar KG. Ligand binding and activation of the secretin receptor, a prototypic family B G protein-coupled receptor. Br J Pharmacol. 2012 May;166(1):18-26. doi: 10.1111/j.1476-5381.2011.01463.x. PMID:21542831 doi:http://dx.doi.org/10.1111/j.1476-5381.2011.01463.x
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 'Lehninger A., Nelson D.N, & Cox M.M. (2008) Lehninger Principles of Biochemistry. W. H. Freeman, fifth edition.'
- ↑ 12.0 12.1 Yang DH, Zhou CH, Liu Q, Wang MW. Landmark studies on the glucagon subfamily of GPCRs: from small molecule modulators to a crystal structure. Acta Pharmacol Sin. 2015 Sep;36(9):1033-42. doi: 10.1038/aps.2015.78. Epub 2015, Aug 17. PMID:26279155 doi:http://dx.doi.org/10.1038/aps.2015.78
- ↑ Ahren B. Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat Rev Drug Discov. 2009 May;8(5):369-85. doi: 10.1038/nrd2782. Epub 2009 Apr, 14. PMID:19365392 doi:http://dx.doi.org/10.1038/nrd2782
- ↑ Xu Y, Xie X. Glucagon receptor mediates calcium signaling by coupling to G alpha q/11 and G alpha i/o in HEK293 cells. J Recept Signal Transduct Res. 2009 Dec;29(6):318-25. doi:, 10.3109/10799890903295150. PMID:19903011 doi:http://dx.doi.org/10.3109/10799890903295150
- ↑ 15.0 15.1 Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ 16.0 16.1 Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859
- ↑ Thomsen J, Kristiansen K, Brunfeldt K, Sundby F. The amino acid sequence of human glucagon. FEBS Lett. 1972 Apr 1;21(3):315-319. PMID:11946536
- ↑ 18.0 18.1 18.2 Weston C, Lu J, Li N, Barkan K, Richards GO, Roberts DJ, Skerry TM, Poyner D, Pardamwar M, Reynolds CA, Dowell SJ, Willars GB, Ladds G. Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2). J Biol Chem. 2015 Sep 18;290(38):23009-22. doi: 10.1074/jbc.M114.624601. Epub, 2015 Jul 21. PMID:26198634 doi:http://dx.doi.org/10.1074/jbc.M114.624601
- ↑ 19.0 19.1 Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, Barwell J, Drmota T, Poyner DR. Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br J Pharmacol. 2013 Feb;168(4):822-34. doi: 10.1111/j.1476-5381.2012.02202.x. PMID:22946657 doi:http://dx.doi.org/10.1111/j.1476-5381.2012.02202.x
- ↑ 20.0 20.1 Lau J, Behrens C, Sidelmann UG, Knudsen LB, Lundt B, Sams C, Ynddal L, Brand CL, Pridal L, Ling A, Kiel D, Plewe M, Shi S, Madsen P. New beta-alanine derivatives are orally available glucagon receptor antagonists. J Med Chem. 2007 Jan 11;50(1):113-28. PMID:17201415 doi:http://dx.doi.org/10.1021/jm058026u