1jqn
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1jqn |SIZE=350|CAPTION= <scene name='initialview01'>1jqn</scene>, resolution 2.35Å | |PDB= 1jqn |SIZE=350|CAPTION= <scene name='initialview01'>1jqn</scene>, resolution 2.35Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= | + | |LIGAND= <scene name='pdbligand=ASP:ASPARTIC+ACID'>ASP</scene>, <scene name='pdbligand=DCO:3,3-DICHLORO-2-PHOSPHONOMETHYL-ACRYLIC+ACID'>DCO</scene>, <scene name='pdbligand=MN:MANGANESE+(II)+ION'>MN</scene> |
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Phosphoenolpyruvate_carboxylase Phosphoenolpyruvate carboxylase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.1.31 4.1.1.31] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Phosphoenolpyruvate_carboxylase Phosphoenolpyruvate carboxylase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.1.31 4.1.1.31] </span> |
|GENE= Escherichia coli ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli]) | |GENE= Escherichia coli ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli]) | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[1fiy|1FIY]], [[1jqo|1jqo]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1jqn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1jqn OCA], [http://www.ebi.ac.uk/pdbsum/1jqn PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1jqn RCSB]</span> | ||
}} | }} | ||
| Line 25: | Line 28: | ||
[[Category: Kai, Y.]] | [[Category: Kai, Y.]] | ||
[[Category: Matsumura, H.]] | [[Category: Matsumura, H.]] | ||
| - | [[Category: ASP]] | ||
| - | [[Category: DCO]] | ||
| - | [[Category: MN]] | ||
[[Category: beta barrel]] | [[Category: beta barrel]] | ||
[[Category: mn2+ and dcdp complex]] | [[Category: mn2+ and dcdp complex]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 21:37:33 2008'' |
Revision as of 18:37, 30 March 2008
| |||||||
| , resolution 2.35Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Gene: | Escherichia coli (Escherichia coli) | ||||||
| Activity: | Phosphoenolpyruvate carboxylase, with EC number 4.1.1.31 | ||||||
| Related: | 1FIY, 1jqo
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
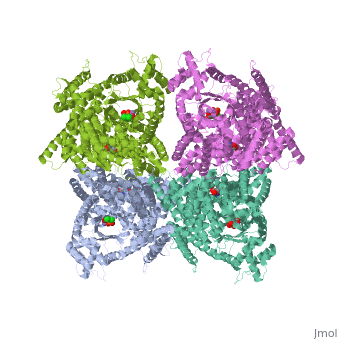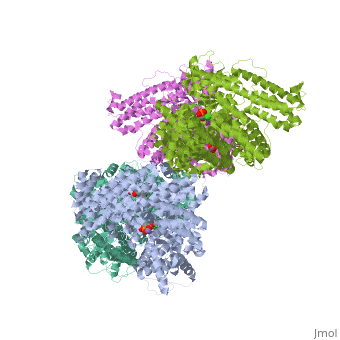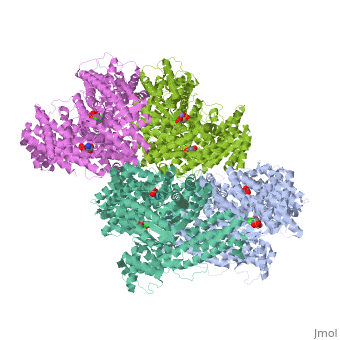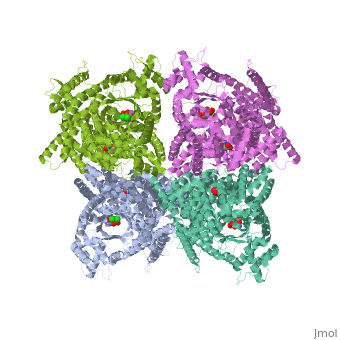
Crystal structure of E.coli phosphoenolpyruvate carboxylase in complex with Mn2+ and DCDP
Overview
Phosphoenolpyruvate carboxylase (PEPC) catalyzes the first step in the fixation of atmospheric CO(2) during C(4) photosynthesis. The crystal structure of C(4) form maize PEPC (ZmPEPC), the first structure of the plant PEPCs, has been determined at 3.0 A resolution. The structure includes a sulfate ion at the plausible binding site of an allosteric activator, glucose 6-phosphate. The crystal structure of E. coli PEPC (EcPEPC) complexed with Mn(2+), phosphoenolpyruvate analog (3,3-dichloro-2-dihydroxyphosphinoylmethyl-2-propenoate), and an allosteric inhibitor, aspartate, has also been determined at 2.35 A resolution. Dynamic movements were found in the ZmPEPC structure, compared with the EcPEPC structure, around two loops near the active site. On the basis of these molecular structures, the mechanisms for the carboxylation reaction and for the allosteric regulation of PEPC are proposed.
About this Structure
1JQN is a Single protein structure of sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases., Matsumura H, Xie Y, Shirakata S, Inoue T, Yoshinaga T, Ueno Y, Izui K, Kai Y, Structure. 2002 Dec;10(12):1721-30. PMID:12467579
Page seeded by OCA on Sun Mar 30 21:37:33 2008