Pyrrolysyl-tRNA synthetase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
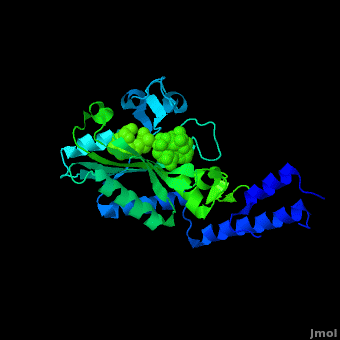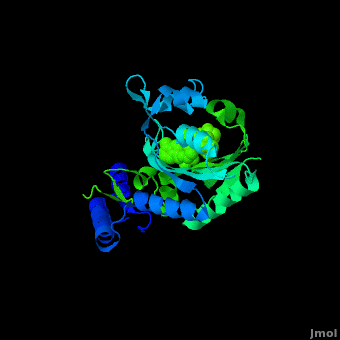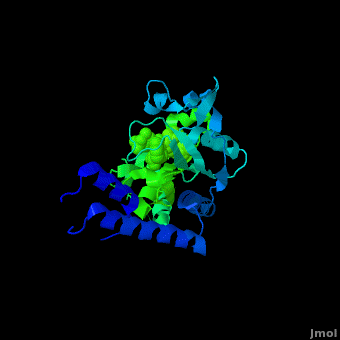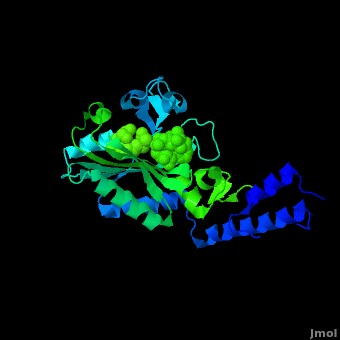
<StructureSection load='2zin' size='450' side='right' scene='Sandbox_166/Scene_2/1' caption='Pyrrolysyl-tRNA synthetase complex with ATP analog and tert-butoxycarbo-lysine (PDB code [[2zin]])'> | <StructureSection load='2zin' size='450' side='right' scene='Sandbox_166/Scene_2/1' caption='Pyrrolysyl-tRNA synthetase complex with ATP analog and tert-butoxycarbo-lysine (PDB code [[2zin]])'> | ||
| - | + | __TOC__ | |
== Function == | == Function == | ||
[[Pyrrolysyl-tRNA synthetase]] (PyIRS) is encoded by the gene pyIS and is found to belong as a part of the group of enzymatic proteins whose role involves the cellular process of tRNA aminoacylation required for protein translation.<ref name="trans"> PMID:17267409 </ref> In particular, PyIRS is required for the activation of the amino acid [[Pyrrolysine]] as it associates with a tRNA generating a specific tRNA<sup>Pyl</sup>, which is then further used to transfer the amino acid to a growing polypeptide.<ref name="pept"> PMID:19022179 </ref> The involvement of PyIRS is carried out due to the anticodon CUA on the suppressor tRNA<sup>Pyl</sup> that is complementary to the UAG codon.<ref name="amber"> PMID:1796745 </ref> <ref>PMID:15314242 </ref> The interesting fact is that this is done by the response of the codon UAG (amber codon) on the mRNA that is normally a stop codon in other organisms. Pyrrolysine (Pyl) is the 22nd existing amino acid genetically encoded in nature that was first discovered as a byproduct contained by the active site of monomethylamine methyltransferase, exclusively from Methanosarcina barkeri (M. barkeri) species.<ref name="pept" /> <ref name="barkeri"> PMID:19118381 </ref> Thus, it is utilized by a variety of organisms that metabolize methylamines for acquiring energy such as methanogenic Archaea of the family Methanosarcinace; along with two known bacterium species<ref name="trans" /> <ref name="barkeri" /> Pyrrolysine’s structural makeup consists of 4-methylpyrroline-5-carboxylate in amide linkage with the N<sup>ϵ</sup> of lysine.<ref name="lysine">PMID:16096277 </ref> This arrangement is comparable to lysine; however, being its derivative it contains an added pyrroline ring that is found to lie situated at the back of the structure.<ref name="lysine" /> | [[Pyrrolysyl-tRNA synthetase]] (PyIRS) is encoded by the gene pyIS and is found to belong as a part of the group of enzymatic proteins whose role involves the cellular process of tRNA aminoacylation required for protein translation.<ref name="trans"> PMID:17267409 </ref> In particular, PyIRS is required for the activation of the amino acid [[Pyrrolysine]] as it associates with a tRNA generating a specific tRNA<sup>Pyl</sup>, which is then further used to transfer the amino acid to a growing polypeptide.<ref name="pept"> PMID:19022179 </ref> The involvement of PyIRS is carried out due to the anticodon CUA on the suppressor tRNA<sup>Pyl</sup> that is complementary to the UAG codon.<ref name="amber"> PMID:1796745 </ref> <ref>PMID:15314242 </ref> The interesting fact is that this is done by the response of the codon UAG (amber codon) on the mRNA that is normally a stop codon in other organisms. Pyrrolysine (Pyl) is the 22nd existing amino acid genetically encoded in nature that was first discovered as a byproduct contained by the active site of monomethylamine methyltransferase, exclusively from Methanosarcina barkeri (M. barkeri) species.<ref name="pept" /> <ref name="barkeri"> PMID:19118381 </ref> Thus, it is utilized by a variety of organisms that metabolize methylamines for acquiring energy such as methanogenic Archaea of the family Methanosarcinace; along with two known bacterium species<ref name="trans" /> <ref name="barkeri" /> Pyrrolysine’s structural makeup consists of 4-methylpyrroline-5-carboxylate in amide linkage with the N<sup>ϵ</sup> of lysine.<ref name="lysine">PMID:16096277 </ref> This arrangement is comparable to lysine; however, being its derivative it contains an added pyrroline ring that is found to lie situated at the back of the structure.<ref name="lysine" /> | ||
Revision as of 08:52, 2 August 2016
| |||||||||||
3D structures of pyrrolysyl-tRNA synthetase
Updated on 02-August-2016
Additional Resources
For Additional information, see: Translation
References
- ↑ 1.0 1.1 Herring S, Ambrogelly A, Polycarpo CR, Soll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35(4):1270-8. Epub 2007 Jan 31. PMID:17267409 doi:10.1093/nar/gkl1151
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol. 2008 Nov 24;15(11):1187-97. PMID:19022179 doi:10.1016/j.chembiol.2008.10.004
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Urbancsek J, Rabe T, Grunwald K, Kiesel L, Papp Z, Runnebaum B. High preovulatory serum luteinizing hormone level is unfavorable to conception. Gynecol Endocrinol. 1991 Dec;5(4):223-33. PMID:1796745
- ↑ Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12450-4. Epub 2004 Aug 16. PMID:15314242 doi:10.1073/pnas.0405362101
- ↑ 5.0 5.1 5.2 Nozawa K, O'Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Soll D, Nureki O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009 Feb 26;457(7233):1163-7. Epub 2008 Dec 31. PMID:19118381 doi:10.1038/nature07611
- ↑ 6.0 6.1 Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J Biol Chem. 2005 Nov 4;280(44):36962-9. Epub 2005 Aug 11. PMID:16096277 doi:10.1074/jbc.M506402200
- ↑ Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006 Dec 11;580(28-29):6695-700. Epub 2006 Nov 20. PMID:17126325 doi:10.1016/j.febslet.2006.11.028
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol. 2008 May 2;378(3):634-52. Epub 2008 Feb 29. PMID:18387634 doi:10.1016/j.jmb.2008.02.045
- ↑ 9.0 9.1 9.2 9.3 Ibba M, Soll D. Genetic code: introducing pyrrolysine. Curr Biol. 2002 Jul 9;12(13):R464-6. PMID:12121639
Proteopedia Page Contributors and Editors (what is this?)
Agatka Jaworski, Michal Harel, Joel L. Sussman, David Canner, Andrea Gorrell, Alexander Berchansky