We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Porphobilinogen synthase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='1h7n' size='450' side='right' caption='Yeast porphobilinogen synthase complex with laevulunic acid and Zn+2 ion (grey) (PDB entry [[1h7n]])' scene=''> | + | <StructureSection load='1h7n' size='450' side='right' caption='Yeast porphobilinogen synthase complex with laevulunic acid and Zn+2 ion (grey) (PDB entry [[1h7n]])' scene='52/526179/Cv/1'> |
== Function == | == Function == | ||
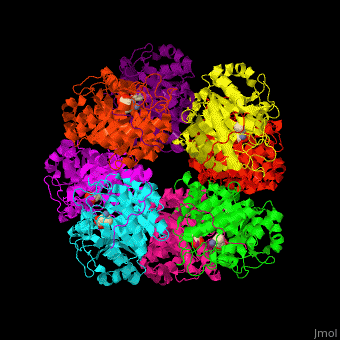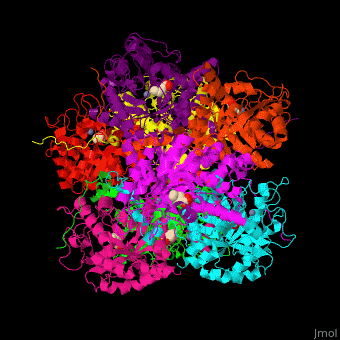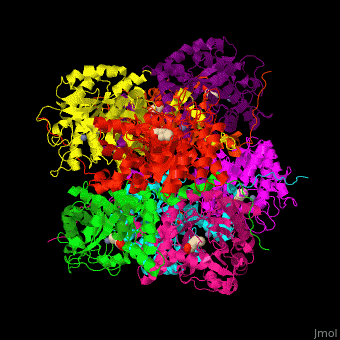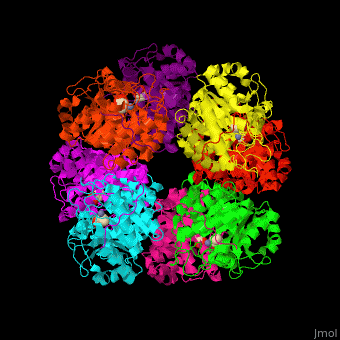
'''Porphobilinogen synthase''' (PBS) catalyzes the formation of porphobilinogen (PBG) from 2 molecules of aminolaevulinic acid (ALA). PBS participates in porphyrin biosynthesis<ref>PMID:15381398</ref>. Levulinic acid (LA) is an inhibitor of PBS. Porphyrin is the precursor of hemes, chlorophyll and vitamin B12. | '''Porphobilinogen synthase''' (PBS) catalyzes the formation of porphobilinogen (PBG) from 2 molecules of aminolaevulinic acid (ALA). PBS participates in porphyrin biosynthesis<ref>PMID:15381398</ref>. Levulinic acid (LA) is an inhibitor of PBS. Porphyrin is the precursor of hemes, chlorophyll and vitamin B12. | ||
| Line 7: | Line 7: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The active site of PBS contains the substrate laevulinic acid and Zn+2 ion<ref>PMID:11545591</ref>. | + | The biological assembly of Yeast porphobilinogen synthase is <scene name='52/526179/Cv/2'>homooctomer</scene>. The active site of PBS contains the substrate laevulinic acid and Zn+2 ion<ref>PMID:11545591</ref>. |
</StructureSection> | </StructureSection> | ||
Revision as of 08:44, 8 August 2016
| |||||||||||
3D structures of porphobilinogen synthase
Updated on 08-August-2016
References
- ↑ Jaffe EK. The porphobilinogen synthase catalyzed reaction mechanism. Bioorg Chem. 2004 Oct;32(5):316-25. PMID:15381398 doi:http://dx.doi.org/10.1016/j.bioorg.2004.05.010
- ↑ Doss M, Muller WA. Acute lead poisoning in inherited porphobilinogen synthase (delta-aminolevulinic acid dehydrase) deficiency. Blut. 1982 Aug;45(2):131-9. PMID:7104498
- ↑ Erskine PT, Newbold R, Brindley AA, Wood SP, Shoolingin-Jordan PM, Warren MJ, Cooper JB. The x-ray structure of yeast 5-aminolaevulinic acid dehydratase complexed with substrate and three inhibitors. J Mol Biol. 2001 Sep 7;312(1):133-41. PMID:11545591 doi:10.1006/jmbi.2001.4947