Poly (ADP-ribose) glycohydrolase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
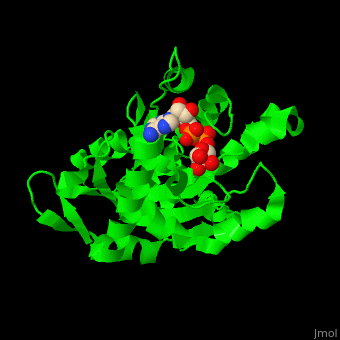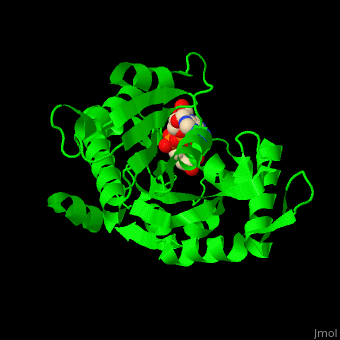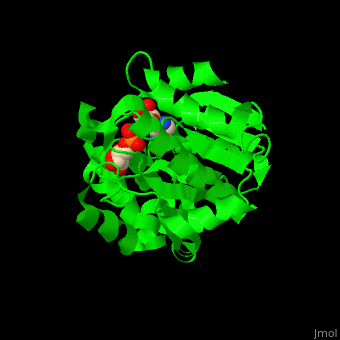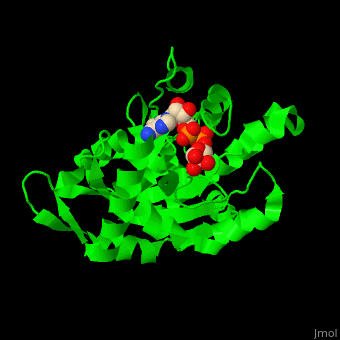
| - | <StructureSection load='3sig' size='450' side='right' caption='Structure of human poly (ADP-ribose) glycohydrolase catalytic domain complex with ADP-ribose | + | <StructureSection load='3sig' size='450' side='right' caption='Structure of human poly (ADP-ribose) glycohydrolase catalytic domain complex with ADP-ribose (PDB code [[3sig]]). ' scene='59/595759/Cv/1'> |
== Function == | == Function == | ||
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The active site of PARG contains two catalytic glutamic acid residues<ref>PMID:21892188</ref>. | + | The <scene name='59/595759/Cv/3'>active site</scene> of PARG contains <scene name='59/595759/Cv/2'>two catalytic glutamic acid residues</scene><ref>PMID:21892188</ref>. Water molecules shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 11:42, 10 August 2016
| |||||||||||
3D Structures of poly (ADP-ribose) glycohydrolase
Updated on 10-August-2016
References
- ↑ Herceg Z, Wang ZQ. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat Res. 2001 Jun 2;477(1-2):97-110. PMID:11376691
- ↑ Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002 Sep;54(3):375-429. PMID:12223530
- ↑ Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011 Sep 4. doi: 10.1038/nature10404. PMID:21892188 doi:10.1038/nature10404