We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) RNA polymerase
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Structural highlights == | == Structural highlights == | ||
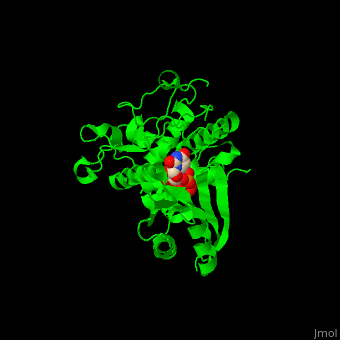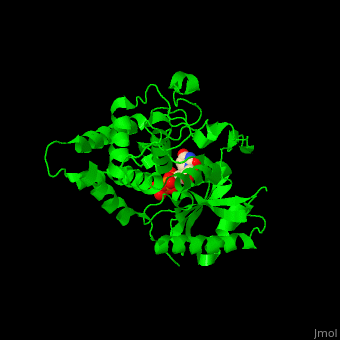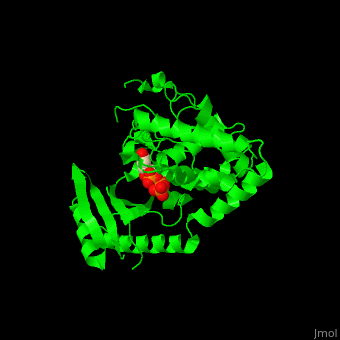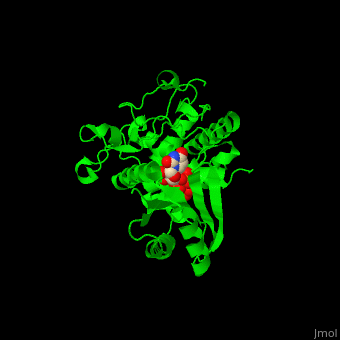
| - | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The active site contains the catalytic triad of aspartic acids in the N-terminal and the His residue whose two conformations select between binding of UTP and ATP to Cid1 <ref>PMID:22751018</ref>. | + | The UTP binding site of Cid1 is between the N-terminal and C-terminal. The <scene name='70/706259/Cv/2'>active site</scene> contains the <scene name='70/706259/Cv/3'>catalytic triad of aspartic acids</scene> (in magenta) in the N-terminal and the <scene name='70/706259/Cv/4'>His residue</scene> (in cyan) whose two conformations select between binding of UTP and ATP to Cid1 <ref>PMID:22751018</ref>. Water molecules shown as red spheres. |
</StructureSection> | </StructureSection> | ||
Revision as of 12:05, 11 August 2016
| |||||||||||
3D Structures of poly(A) RNA polymerase protein Cid1
Updated on 11-August-2016
4e7x, 4e8f – Cid1
4ud4, 4ud5 – Cid1 (mutant)
4e80 – Cid1 + UTP
4fh3 – Cid1 residues 33-377
4ep7, 4fh5, 4fhp – Cid1 residues 30-377 + UTP
4fhv – Cid1 residues 30-377 + CTP
4fhw – Cid1 residues 30-377 + GTP
4fhy – Cid1 residues 30-377 + ATP
4fhx – Cid1 residues 30-377 (mutant) + ATP
4nkt – Cid1 residues 30-377 + UMPNPP
4nku – Cid1 residues 40-377 (mutant) + dinucleotide
References
- ↑ Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006 Oct 15;23(13):991-1000. PMID:17072891 doi:http://dx.doi.org/10.1002/yea.1408
- ↑ Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol. 2012 Jul 1. doi: 10.1038/nsmb.2329. PMID:22751018 doi:10.1038/nsmb.2329
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky