We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
S100 protein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='' size='450' side='right' scene='Journal:JBIC:18/Cv/1' caption=''> | <StructureSection load='' size='450' side='right' scene='Journal:JBIC:18/Cv/1' caption=''> | ||
| - | + | == Function == | |
| - | '''S100 proteins''' are calcium-binding proteins (CBP) found in vertebrates and contain 2 helix-loop-helix calcium binding sites. S100 name is derived from their being 100% soluble in ammonium sulfate. There are at least 21 S100 proteins. Most S100 proteins undergo conformational change upon calcium binding.<br /> | + | '''S100 proteins''' are calcium-binding proteins (CBP) found in vertebrates and contain 2 helix-loop-helix calcium binding sites. S100 name is derived from their being 100% soluble in ammonium sulfate. There are at least 21 S100 proteins. Most S100 proteins undergo conformational change upon calcium binding<ref>PMID:22834835</ref>.<br /> |
'''S100-A1''' is a regulator of myocardial contractility.<br /> | '''S100-A1''' is a regulator of myocardial contractility.<br /> | ||
'''S100-A2''' may have a tumor suppression function.<br /> | '''S100-A2''' may have a tumor suppression function.<br /> | ||
'''S100-A4 and S100-A11''' may function in motility, invasion and tubulin polymerization.<br /> | '''S100-A4 and S100-A11''' may function in motility, invasion and tubulin polymerization.<br /> | ||
'''S100-A6''' may function in calcium-dependent insulin release.<br /> | '''S100-A6''' may function in calcium-dependent insulin release.<br /> | ||
| - | '''S100-A7''' is an antimicrobial peptide mainly against E. coli.<br /> | + | '''S100-A7''' is an antimicrobial peptide mainly against ''E. coli''.<br /> |
'''S100-A8''' combined with '''S100-A9''' ('''calprotectin''') may function in the inhibition of casein kinase and regulate inflammatory processes.<br /> | '''S100-A8''' combined with '''S100-A9''' ('''calprotectin''') may function in the inhibition of casein kinase and regulate inflammatory processes.<br /> | ||
'''S100-A10''' is implicated in the transport of neurotransmitters.<br /> | '''S100-A10''' is implicated in the transport of neurotransmitters.<br /> | ||
| Line 14: | Line 14: | ||
'''S100G''' is vitamin-D dependent calcium-binding protein and is called calbindin D9k and calbindin D28k depending on its molecular weight. S100G mediates the transport of calcium across enterocytes. | '''S100G''' is vitamin-D dependent calcium-binding protein and is called calbindin D9k and calbindin D28k depending on its molecular weight. S100G mediates the transport of calcium across enterocytes. | ||
| - | === Solution structure and dynamics of human S100A14 <ref>pmid 23197251 </ref> | + | == Relevance == |
| + | S100-A2, S100-A3, S100-A5, S100-A7, S100-A8, S100-A9, S100-A14, S100-A15, S100-A16 and S100-P are over expressed on bladder cancer while S100-A1, S100-A4 and S100-B are under expressed<ref>PMID:17970044</ref>. S100-A12 is the most significant marker for psoriasis disease activity<ref>PMID:26333514</ref>. Various S100 proteins change this expression in skin and sweat glands tumors<ref>PMID:23623205</ref>. | ||
| + | |||
| + | == Solution structure and dynamics of human S100A14 <ref>pmid 23197251 </ref>== | ||
Human <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is a member of the EF-hand calcium-binding protein family that has only recently been described in its functional and pathological properties. The protein is overexpressed in a variety of tumor cells and it has been shown to trigger receptor for advanced glycation end products (RAGE)-dependent signaling in cell cultures. The protein has a high degree of sequence homology with S100A13 although it exhibits a negligible affinity for calcium(II) ions and undergoes aggregation and precipitation in the presence of zinc(II) or copper(II) ions. The lack of two ligands in the canonical EF-hand calcium(II)-binding site explains the negligible affinity for calcium(II) in solution, while the exposed cysteines and histidine account for the observed precipitation in the presence of zinc(II) or copper(II) ions. <scene name='Journal:JBIC:18/Cv/3'>Some residues, responsible for the binding of calcium(II)</scene> in S100A13 as well as in other S100 proteins, are mutated in S100A14. The solution structure of <scene name='Journal:JBIC:18/Cv/5'>homodimeric S100A14</scene> (<span style="color:mediumslateblue;background-color:black;font-weight:bold;">monomer A is in mediumslateblue</span> and <span style="color:deeppink;background-color:black;font-weight:bold;">monomer B is in deeppink</span>) in the apo state at physiological temperature shows close structural similarities with the C-terminal domain of the myosin light chains and folds in a ‘semi-open’ conformation, with helices III and IV showing a X-shaped offset arrangement and helices II and III showing a high number of interactions creating a V-shaped arrangement. | Human <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is a member of the EF-hand calcium-binding protein family that has only recently been described in its functional and pathological properties. The protein is overexpressed in a variety of tumor cells and it has been shown to trigger receptor for advanced glycation end products (RAGE)-dependent signaling in cell cultures. The protein has a high degree of sequence homology with S100A13 although it exhibits a negligible affinity for calcium(II) ions and undergoes aggregation and precipitation in the presence of zinc(II) or copper(II) ions. The lack of two ligands in the canonical EF-hand calcium(II)-binding site explains the negligible affinity for calcium(II) in solution, while the exposed cysteines and histidine account for the observed precipitation in the presence of zinc(II) or copper(II) ions. <scene name='Journal:JBIC:18/Cv/3'>Some residues, responsible for the binding of calcium(II)</scene> in S100A13 as well as in other S100 proteins, are mutated in S100A14. The solution structure of <scene name='Journal:JBIC:18/Cv/5'>homodimeric S100A14</scene> (<span style="color:mediumslateblue;background-color:black;font-weight:bold;">monomer A is in mediumslateblue</span> and <span style="color:deeppink;background-color:black;font-weight:bold;">monomer B is in deeppink</span>) in the apo state at physiological temperature shows close structural similarities with the C-terminal domain of the myosin light chains and folds in a ‘semi-open’ conformation, with helices III and IV showing a X-shaped offset arrangement and helices II and III showing a high number of interactions creating a V-shaped arrangement. | ||
| Line 23: | Line 26: | ||
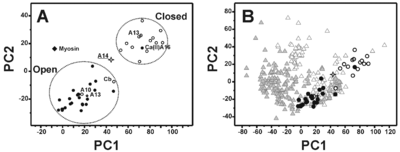
The most remarkable feature of this structural conformation involves the packing of the helices that is reduced with respect to the ‘closed’ structures of the S100 proteins but is still sizably larger than the corresponding ‘open’ structures. At the same time, the analysis of the electrostatic potential surface suggests that the <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is <scene name='Journal:JBIC:18/Cv/7'>permanently activated and it is not calcium(II) regulated</scene>. | The most remarkable feature of this structural conformation involves the packing of the helices that is reduced with respect to the ‘closed’ structures of the S100 proteins but is still sizably larger than the corresponding ‘open’ structures. At the same time, the analysis of the electrostatic potential surface suggests that the <scene name='Journal:JBIC:18/Cv/2'>S100A14</scene> is <scene name='Journal:JBIC:18/Cv/7'>permanently activated and it is not calcium(II) regulated</scene>. | ||
| - | + | == Structural characterization of human S100A16, a low-affinity calcium binder <ref >DOI 10.1007/s00775-010-0721-3</ref>== | |
S100A16 is a special member of the S100 class of calcium binding proteins, because it <scene name='Journal:JBIC:3/As/12'>performs a conformational change upon calcium(II) binding</scene> much smaller than experienced by most S100 proteins. This was observed after determination of the solution structures of apo and <scene name='Journal:JBIC:3/Dual_binding_calcium/3'>calcium(II)-bound S100A16</scene> and the <scene name='Journal:JBIC:3/Crysal/2'>crystal structure of apo S100A16</scene>. The likely reason for minimal conformational change <scene name='Journal:JBIC:3/Calcium_binding_start/7'>in S100A16</scene> is the lower calcium binding affinity and stronger <scene name='Journal:JBIC:3/Hydrophobic_interactions_2/3'>hydrophobic interaction</scene> between <scene name='Journal:JBIC:3/Please_work/3'>helix III and IV present in this protein </scene> with respect to other S100 proteins. Another characteristic of <scene name='Journal:JBIC:3/Opening/3'>S100A16</scene> is that the helix IV has the same length in <scene name='Journal:JBIC:3/25_residue_long_apo/3'>both apo</scene> and <scene name='Journal:JBIC:3/25_residue_calclium_bound/3'>calcium(II) forms</scene> because of <scene name='Journal:JBIC:3/Motif_good/5'>the presence of a Gly-Gly-Ile-Thr-Gly-Pro sequence motif</scene> in helix IV. Based on the available structures of S100 members, we analyzed and summarized all their conformational changes due to calcium(II) binding by a principal component analysis. <scene name='Journal:JBIC:3/Calcium_binding_start/7'>Calcium binding</scene> was proved by both NMR titration and Isothermal Titration Calorimetry (ITC) experiments. Even if the <scene name='Journal:JBIC:3/Binding_calcium_glu/2'>important Glu residue</scene> in the last position of first EF-hand calcium binding loop <scene name='Journal:JBIC:3/Binding_calcium/13'>is missing</scene>, these experimental data indicated that S100A16 can still bind one calcium(II) ion in such loop. NMR relaxation <scene name='Journal:JBIC:3/Flexible_broadwide/4'>studies showed that the first calcium binding loop and the beginning of the second helix</scene> are the most <scene name='Journal:JBIC:3/Flexible_broad/3'>flexible regions in both the apo and calcium(II)-bound S100A16</scene>. Although the biological function of S100A16 is still unclear yet, these structural and dynamic properties can provide useful information for further functional studies. | S100A16 is a special member of the S100 class of calcium binding proteins, because it <scene name='Journal:JBIC:3/As/12'>performs a conformational change upon calcium(II) binding</scene> much smaller than experienced by most S100 proteins. This was observed after determination of the solution structures of apo and <scene name='Journal:JBIC:3/Dual_binding_calcium/3'>calcium(II)-bound S100A16</scene> and the <scene name='Journal:JBIC:3/Crysal/2'>crystal structure of apo S100A16</scene>. The likely reason for minimal conformational change <scene name='Journal:JBIC:3/Calcium_binding_start/7'>in S100A16</scene> is the lower calcium binding affinity and stronger <scene name='Journal:JBIC:3/Hydrophobic_interactions_2/3'>hydrophobic interaction</scene> between <scene name='Journal:JBIC:3/Please_work/3'>helix III and IV present in this protein </scene> with respect to other S100 proteins. Another characteristic of <scene name='Journal:JBIC:3/Opening/3'>S100A16</scene> is that the helix IV has the same length in <scene name='Journal:JBIC:3/25_residue_long_apo/3'>both apo</scene> and <scene name='Journal:JBIC:3/25_residue_calclium_bound/3'>calcium(II) forms</scene> because of <scene name='Journal:JBIC:3/Motif_good/5'>the presence of a Gly-Gly-Ile-Thr-Gly-Pro sequence motif</scene> in helix IV. Based on the available structures of S100 members, we analyzed and summarized all their conformational changes due to calcium(II) binding by a principal component analysis. <scene name='Journal:JBIC:3/Calcium_binding_start/7'>Calcium binding</scene> was proved by both NMR titration and Isothermal Titration Calorimetry (ITC) experiments. Even if the <scene name='Journal:JBIC:3/Binding_calcium_glu/2'>important Glu residue</scene> in the last position of first EF-hand calcium binding loop <scene name='Journal:JBIC:3/Binding_calcium/13'>is missing</scene>, these experimental data indicated that S100A16 can still bind one calcium(II) ion in such loop. NMR relaxation <scene name='Journal:JBIC:3/Flexible_broadwide/4'>studies showed that the first calcium binding loop and the beginning of the second helix</scene> are the most <scene name='Journal:JBIC:3/Flexible_broad/3'>flexible regions in both the apo and calcium(II)-bound S100A16</scene>. Although the biological function of S100A16 is still unclear yet, these structural and dynamic properties can provide useful information for further functional studies. | ||
| Line 201: | Line 204: | ||
**[[2y5i]] – CBP + Ca - zebrafish <br /> | **[[2y5i]] – CBP + Ca - zebrafish <br /> | ||
}} | }} | ||
| + | == References == | ||
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
<references/> | <references/> | ||
Revision as of 07:04, 21 August 2016
| |||||||||||
3D Structures of S100 proteins
Updated on 21-August-2016
References
- ↑ Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med. 2013 Jan;13(1):24-57. PMID:22834835
- ↑ Yao R, Lopez-Beltran A, Maclennan GT, Montironi R, Eble JN, Cheng L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res. 2007 Sep-Oct;27(5A):3051-8. PMID:17970044
- ↑ Wilsmann-Theis D, Wagenpfeil J, Holzinger D, Roth J, Koch S, Schnautz S, Bieber T, Wenzel J. Among the S100 proteins, S100A12 is the most significant marker for psoriasis disease activity. J Eur Acad Dermatol Venereol. 2016 Jul;30(7):1165-70. doi: 10.1111/jdv.13269., Epub 2015 Sep 2. PMID:26333514 doi:http://dx.doi.org/10.1111/jdv.13269
- ↑ Zhu L, Okano S, Takahara M, Chiba T, Tu Y, Oda Y, Furue M. Expression of S100 protein family members in normal skin and sweat gland tumors. J Dermatol Sci. 2013 Jun;70(3):211-9. doi: 10.1016/j.jdermsci.2013.03.002. Epub, 2013 Mar 16. PMID:23623205 doi:http://dx.doi.org/10.1016/j.jdermsci.2013.03.002
- ↑ Bertini I, Borsi V, Cerofolini L, Das Gupta S, Fragai M, Luchinat C. Solution structure and dynamics of human S100A14. J Biol Inorg Chem. 2012 Nov 30. PMID:23197251 doi:10.1007/s00775-012-0963-3
- ↑ Babini E, Bertini I, Borsi V, Calderone V, Hu X, Luchinat C, Parigi G. Structural characterization of human S100A16, a low-affinity calcium binder. J Biol Inorg Chem. 2010 Nov 3. PMID:21046186 doi:10.1007/s00775-010-0721-3