We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SAM-dependent methyltransferase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
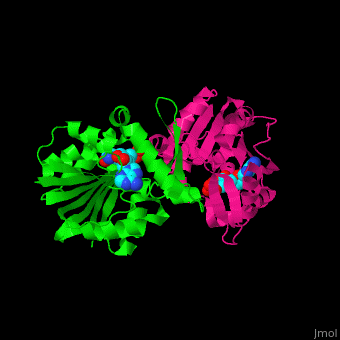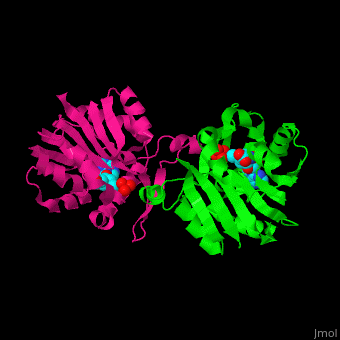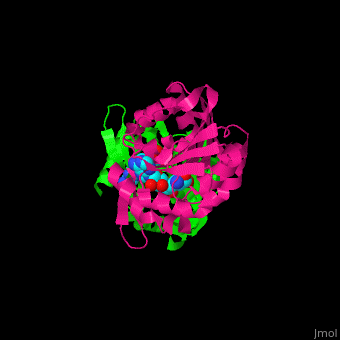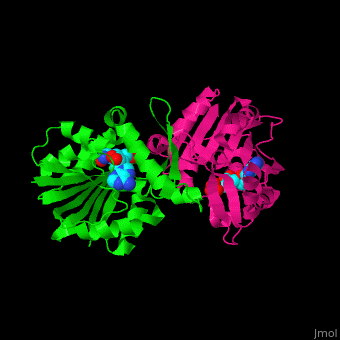
{{STRUCTURE_3h2b| PDB=3h2b | SIZE=400| SCENE= |right|CAPTION=SAM-dependent methyltransferase dimer complex with S-adenosyl-L-homocysteine and pyrophosphate, [[3h2b]] }} | {{STRUCTURE_3h2b| PDB=3h2b | SIZE=400| SCENE= |right|CAPTION=SAM-dependent methyltransferase dimer complex with S-adenosyl-L-homocysteine and pyrophosphate, [[3h2b]] }} | ||
== Function == | == Function == | ||
| - | '''SAM-dependent methyltransferase''' (SDM) utilizes the methyl donor S-adenosyl-L-methionine (SAM) as a cofactor to methylate proteins, small molecules, lipids and nucleic acids. SAM forms S-adenosyl-L-homocysteine (SAH) upon demethylation. About 120 members of the SDM family have been identified. They differ in their substrate specificity and the atom targeted for methylation (N, O, C, S)<ref>PMID:23180741</ref>. | + | '''SAM-dependent methyltransferase''' (SDM) utilizes the methyl donor S-adenosyl-L-methionine (SAM) as a cofactor to methylate proteins, small molecules, lipids and nucleic acids. SAM forms S-adenosyl-L-homocysteine (SAH) upon demethylation. About 120 members of the SDM family have been identified. They differ in their substrate specificity and the atom targeted for methylation (N, O, C, S)<ref>PMID:23180741</ref>. |
| + | <StructureSection load='1af7' size='350' side='right' scene='47/479962/Cv/1' caption='Chemotaxis receptor methyltransferase CheR complex with S-adenosyl-L-homocysteine, [[1af7]]'> | ||
| + | |||
| + | For '''Chemotaxis receptor methyltransferase CheR''' see details in [[Molecular Playgroun]d/CheR]].<ref>PMID:9628482</ref> | ||
| + | *<scene name='47/479962/Cv/2'>S-adenosyl-L-homocysteine binding site</scene> of Chemotaxis receptor methyltransferase CheR ([[1af7]]).<ref>PMID:9115443</ref> | ||
| + | *SEE ALSO [[Chemotaxis protein]]. | ||
| + | </StructureSection> | ||
== Structural highlights == | == Structural highlights == | ||
| Line 44: | Line 50: | ||
**[[3h2b]] – CgSDM + pyrophosphate + SAH<br /> | **[[3h2b]] – CgSDM + pyrophosphate + SAH<br /> | ||
**[[5bp7]] – SlSDM + SAH – ''Geobacter sulfurreducens''<br /> | **[[5bp7]] – SlSDM + SAH – ''Geobacter sulfurreducens''<br /> | ||
| + | |||
| + | <StructureSection load='1af7' size='350' side='right' scene='47/479962/Cv/1' caption='Chemotaxis receptor methyltransferase CheR complex with S-adenosyl-L-homocysteine, [[1af7]]'> | ||
| + | |||
| + | For '''Chemotaxis receptor methyltransferase CheR''' see details in [[Molecular Playground/CheR]].<ref>PMID:9628482</ref> | ||
| + | *<scene name='47/479962/Cv/2'>S-adenosyl-L-homocysteine binding site</scene> of Chemotaxis receptor methyltransferase CheR ([[1af7]]).<ref>PMID:9115443</ref> | ||
| + | *SEE ALSO [[Chemotaxis protein]]. | ||
| + | </StructureSection> | ||
| + | *Chemotaxis receptor methyltransferase (CheR) | ||
| + | |||
| + | **[[1af7]] – StCheR – ''Salmonella typhimurium''<br /> | ||
| + | **[[1bc5]] – StCheR + chemotaxis receptor peptide | ||
}} | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 07:56, 23 August 2016
Contents |
Function
SAM-dependent methyltransferase (SDM) utilizes the methyl donor S-adenosyl-L-methionine (SAM) as a cofactor to methylate proteins, small molecules, lipids and nucleic acids. SAM forms S-adenosyl-L-homocysteine (SAH) upon demethylation. About 120 members of the SDM family have been identified. They differ in their substrate specificity and the atom targeted for methylation (N, O, C, S)[1].
| |||||||||||
Structural highlights
The core of the SDM fold contains alternating β strands and α helices.
3D structures of SAM-dependent methyltrasferase
Updated on 23-August-2016
| |||||||||||
References
- ↑ Struck AW, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem. 2012 Dec 21;13(18):2642-55. doi: 10.1002/cbic.201200556. Epub 2012, Nov 23. PMID:23180741 doi:http://dx.doi.org/10.1002/cbic.201200556
- ↑ Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998 Jun;5(6):446-50. PMID:9628482
- ↑ Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997 Apr 15;5(4):545-58. PMID:9115443
- ↑ Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998 Jun;5(6):446-50. PMID:9628482
- ↑ Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997 Apr 15;5(4):545-58. PMID:9115443