We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Penicillin-binding protein
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
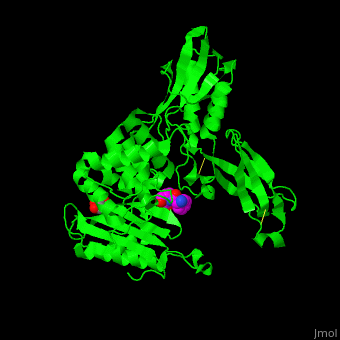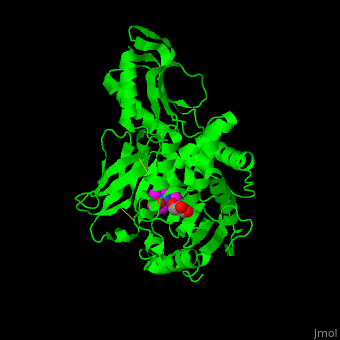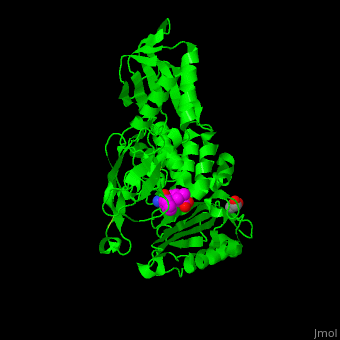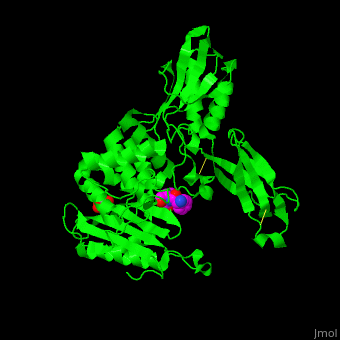
| - | ''E. coli'' PBP structure shows a distinct 3 domain structure. The active site contains the covalently bonded Ser62<ref>PMID:16411754</ref>. | + | ''E. coli'' PBP structure shows a distinct 3 domain structure. The <scene name='47/478544/Cv/2'>active site</scene> contains the <scene name='47/478544/Cv/3'>covalently bonded Ser62</scene><ref>PMID:16411754</ref>. |
</StructureSection> | </StructureSection> | ||
==3D structures of penicillin-binding protein== | ==3D structures of penicillin-binding protein== | ||
Revision as of 10:12, 18 September 2016
| |||||||||||
3D structures of penicillin-binding protein
Updated on 18-September-2016
References
- ↑ Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999-3003. PMID:1103132
- ↑ Beadle BM, Nicholas RA, Shoichet BK. Interaction energies between beta-lactam antibiotics and E. coli penicillin-binding protein 5 by reversible thermal denaturation. Protein Sci. 2001 Jun;10(6):1254-9. PMID:11369864 doi:http://dx.doi.org/10.1110/ps.52001
- ↑ Kishida H, Unzai S, Roper DI, Lloyd A, Park SY, Tame JR. Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics. Biochemistry. 2006 Jan 24;45(3):783-92. PMID:16411754 doi:10.1021/bi051533t