1pvo
From Proteopedia
| Line 4: | Line 4: | ||
|PDB= 1pvo |SIZE=350|CAPTION= <scene name='initialview01'>1pvo</scene>, resolution 3.00Å | |PDB= 1pvo |SIZE=350|CAPTION= <scene name='initialview01'>1pvo</scene>, resolution 3.00Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand=ANP:PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER'>ANP</scene> | + | |LIGAND= <scene name='pdbligand=ANP:PHOSPHOAMINOPHOSPHONIC+ACID-ADENYLATE+ESTER'>ANP</scene>, <scene name='pdbligand=C:CYTIDINE-5'-MONOPHOSPHATE'>C</scene>, <scene name='pdbligand=U:URIDINE-5'-MONOPHOSPHATE'>U</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= RHO ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli]) | |GENE= RHO ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli]) | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY=[[1pv4|1PV4]] | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1pvo FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1pvo OCA], [http://www.ebi.ac.uk/pdbsum/1pvo PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1pvo RCSB]</span> | ||
}} | }} | ||
| Line 24: | Line 27: | ||
[[Category: Berger, J M.]] | [[Category: Berger, J M.]] | ||
[[Category: Skordalakes, E.]] | [[Category: Skordalakes, E.]] | ||
| - | [[Category: ANP]] | ||
[[Category: rho-anppnp-ssrna complex]] | [[Category: rho-anppnp-ssrna complex]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 23:05:09 2008'' |
Revision as of 20:05, 30 March 2008
| |||||||
| , resolution 3.00Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Gene: | RHO (Escherichia coli) | ||||||
| Related: | 1PV4
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
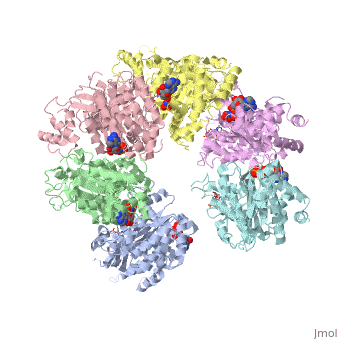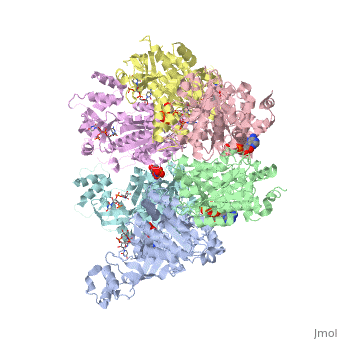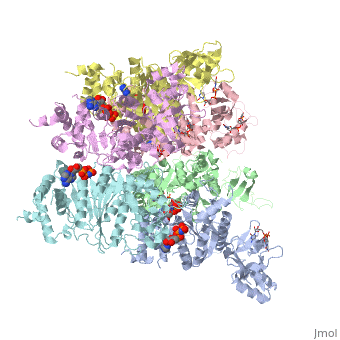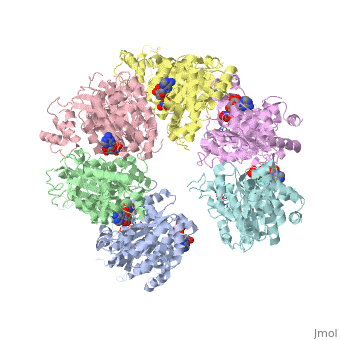
X-ray crystal structure of Rho transcription termination factor in complex with ssRNA substrate and ANPPNP
Overview
In bacteria, one of the major transcriptional termination mechanisms requires a RNA/DNA helicase known as the Rho factor. We have determined two structures of Rho complexed with nucleic acid recognition site mimics in both free and nucleotide bound states to 3.0 A resolution. Both structures show that Rho forms a hexameric ring in which two RNA binding sites--a primary one responsible for target mRNA recognition and a secondary one required for mRNA translocation and unwinding--point toward the center of the ring. Rather than forming a closed ring, the Rho hexamer is split open, resembling a "lock washer" in its global architecture. The distance between subunits at the opening is sufficiently wide (12 A) to accommodate single-stranded RNA. This open configuration most likely resembles a state poised to load onto mRNA and suggests how related ring-shaped enzymes may be breached to bind nucleic acids.
About this Structure
1PVO is a Single protein structure of sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading., Skordalakes E, Berger JM, Cell. 2003 Jul 11;114(1):135-46. PMID:12859904
Page seeded by OCA on Sun Mar 30 23:05:09 2008