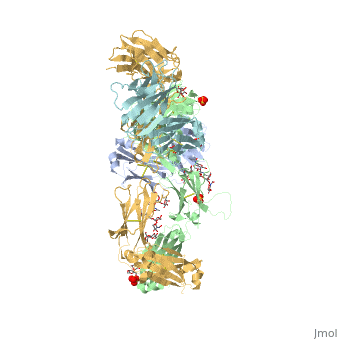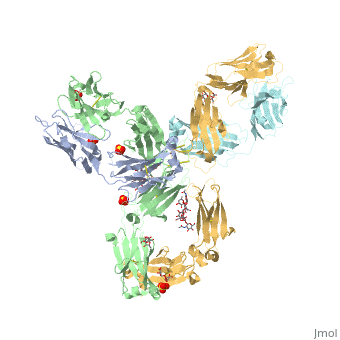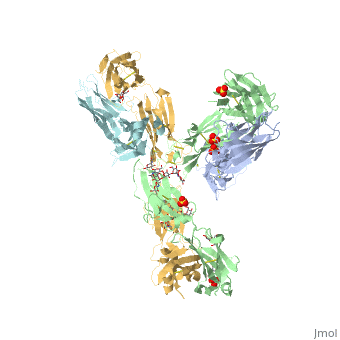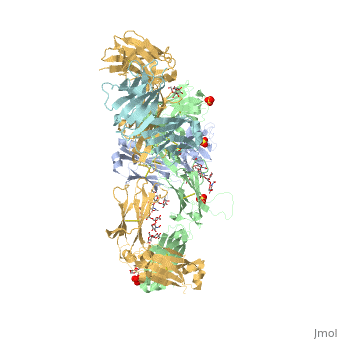We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox454
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Structure and Function == | == Structure and Function == | ||
| - | Pembrolizumab, or Keytruda, is an immunoglobulin G4 (IgG4)-kappa humanized monoclonal antibody against the programmed cell death-1 (PD-1) receptor. It is a very compact molecule with an asymmetrical Y-shape. The short compact hinge region inflicts constraints on the molecule that creates the abnormal crystallizable tail region (Fc domain) compared to other immunoglobulin G (IgG) proteins. The Fc domain is <scene name='74/745945/Glycosylation/1'>glycosylated</scene> at both CH2 domains on each chain and one of them is distinctively rotated 120° compared to other similar structures, making the glycan chain more solvent accessible and facing the solvent. IgG4s have a unique function where they form dynamic bispecific antibodies by exchanging half-molecules (one heavy chain/light chain pair) among themselves, called Fab-arm exchange. This makes the molecule particularly unstable and unpredictable as a treatment, but can be conquered by introducing a serine-to-proline mutation at <scene name='74/745945/Pro228/1'>amino acid 228</scene>, which prevents Fab-arm exchange and stabilizes the molecule <ref>DOI:10.1080/17425255.2016.1216976</ref>. | + | Pembrolizumab, or Keytruda, is an immunoglobulin G4 (IgG4)-kappa humanized monoclonal antibody against the programmed cell death-1 (PD-1) receptor. It is a very compact molecule with an asymmetrical Y-shape. The short compact hinge region inflicts constraints on the molecule that creates the abnormal crystallizable tail region (Fc domain) compared to other immunoglobulin G (IgG) proteins. The Fc domain is <scene name='74/745945/Glycosylation/1'>glycosylated</scene> at both CH2 domains on each chain and one of them is distinctively rotated 120° compared to other similar structures, making the glycan chain more solvent accessible and facing the solvent. IgG4s have a unique function where they form dynamic bispecific antibodies by exchanging half-molecules (one heavy chain/light chain pair) among themselves, called Fab-arm exchange. This makes the molecule particularly unstable and unpredictable as a treatment, but can be conquered by introducing a serine-to-proline mutation at <scene name='74/745945/Pro228/1'>amino acid 228</scene>, which prevents Fab-arm exchange and stabilizes the molecule <ref>DOI:10.1080/17425255.2016.1216976</ref>. Pembrolizumab contains an Fv fragment (PemFv) and a Fab fragment (PemFab). The Fv fragment is the variable region of the molecule where binding occurs whereas the Fab fragment constitutes the whole molecule. |
== Pembrolizumab/PD-1 Interaction == | == Pembrolizumab/PD-1 Interaction == | ||
| Line 14: | Line 14: | ||
== PemFv/PD-1 Interaction == | == PemFv/PD-1 Interaction == | ||
| - | The Fv fragment of pembrolizumab | + | The Fv fragment of pembrolizumab can form a complex with the extracellular domain (ECD) of PD-1. Both PemFv and PD-1ECD contain interchain disulfide bonds. PemFv interacts predominantly in the major groove of PD-1, which is formed on one surface by the CC’FG antiparallel β−sheet and the BC, C’D, and FG loops. There are 15 direct hydrogen bonds between the residues, 15 water-mediated hydrogen bonds, 2 salt bridges, and many hydrophobic interactions. A very large solvent-accessible surface area of 1,137Å2 is buried on PD-1ECD due to the convoluted interaction. There are a total of 26 PD-1ECD residues involved in the interaction with PemFv, with residues in loop C’D (Pro84 to Gly90) and strand C’ (Gln75 to Lys 78) playing a major role. These key components of PD-1 mainly form interactions through salt bridges and hydrogen bonds with CRD-L3, CDR-H1, CDR-H2, CDR-H3 of pembrolizumab. It is beleived that the sugar chains of PD-1 have no physical contact with pembrolizumab due to the N-linked glycosylated residues (Asn49, Asn58, Asn74, and Asn116) being located away from the interaface <ref name="horita" />. |
== PD-L1/PD-1 Interaction == | == PD-L1/PD-1 Interaction == | ||
Revision as of 15:46, 16 November 2016
Pembrolizumab
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976
- ↑ 4.0 4.1 4.2 4.3 Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Rajakulendran T, Adam DN. Spotlight on pembrolizumab in the treatment of advanced melanoma. Drug Des Devel Ther. 2015 Jun 4;9:2883-6. doi: 10.2147/DDDT.S78036. eCollection, 2015. PMID:26082618 doi:http://dx.doi.org/10.2147/DDDT.S78036
- ↑ Deeks ED. Pembrolizumab: A Review in Advanced Melanoma. Drugs. 2016 Mar;76(3):375-86. doi: 10.1007/s40265-016-0543-x. PMID:26846323 doi:http://dx.doi.org/10.1007/s40265-016-0543-x
- ↑ Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976