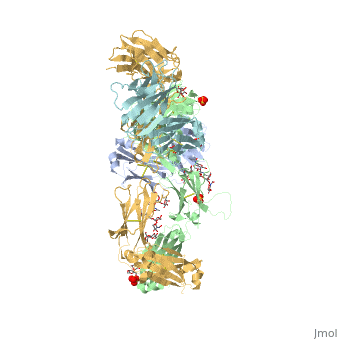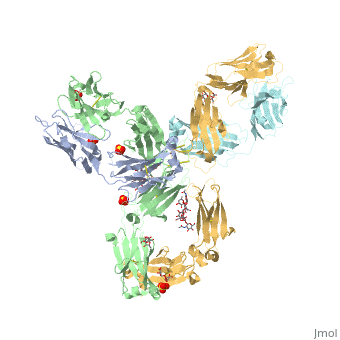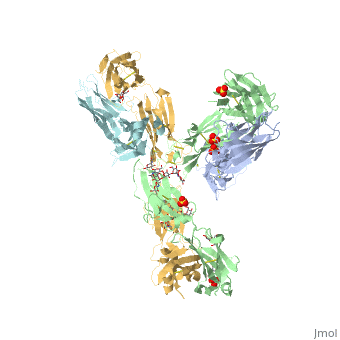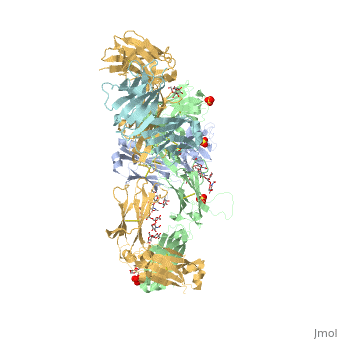Sandbox454
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
== PD-L1/PD-1 Interaction == | == PD-L1/PD-1 Interaction == | ||
| - | The complex formed when protein-derived ligand, PD-L1, interacts with the inhibitory receptor, PD-1, suppresses immune responses | + | The complex formed when protein-derived ligand, PD-L1, interacts with the inhibitory receptor, PD-1, suppresses immune responses against autoantigens and helps in peripheral immune tolerance. However, when tumors over express PD-L1, the interaction with PD-1 inhibits T-lymphocyte proliferation, release of cytokines, and cytotoxicity, exhausting tumor-specific T-cells. There are a total of 12 PD-1ECD residues that are involved in forming the complex with the N-terminal half of PD-L1ECD (PD-L1ECD-N). Nine hydrogen bonds, 3 water-mediated hydrogen bonds, 2 salt bridges, and numerous hydrophobic interactions make up the PD-1ECD/PD-L1ECD-N interaction.The CC’FG sheet within both proteins is the main interaction point. A hydrophobic surface patch is formed when the PD-1ECD is in complex with PD-L1ECD-N. The PD-1ECD residues involved include Val64, Tyr68, Ile126, Leu128, Ala132 and Ile134. Numerous hydrophilic amino acids that encircle PD-L1ECD-N form salt bridges and hydrogen bonds with Asn66, Tyr68, Gln75, Thr76, Asp77, Lys78, Ala132 and Glu136 of PD-1ECD <ref name="horita" />. |
== Disease in Humans - Cancer == | == Disease in Humans - Cancer == | ||
Revision as of 02:58, 17 November 2016
Pembrolizumab/Keytruda
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976
- ↑ 4.0 4.1 4.2 4.3 Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Rajakulendran T, Adam DN. Spotlight on pembrolizumab in the treatment of advanced melanoma. Drug Des Devel Ther. 2015 Jun 4;9:2883-6. doi: 10.2147/DDDT.S78036. eCollection, 2015. PMID:26082618 doi:http://dx.doi.org/10.2147/DDDT.S78036
- ↑ Deeks ED. Pembrolizumab: A Review in Advanced Melanoma. Drugs. 2016 Mar;76(3):375-86. doi: 10.1007/s40265-016-0543-x. PMID:26846323 doi:http://dx.doi.org/10.1007/s40265-016-0543-x