We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Zepatier
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
== Structure and Mechanism of Grazoprevir == | == Structure and Mechanism of Grazoprevir == | ||
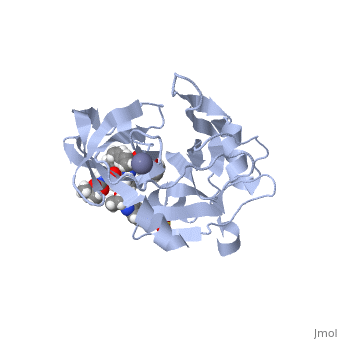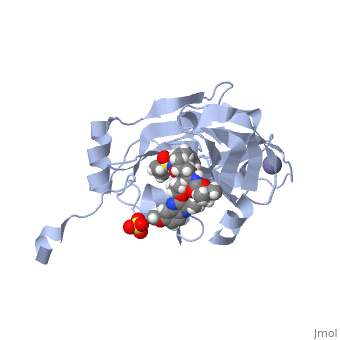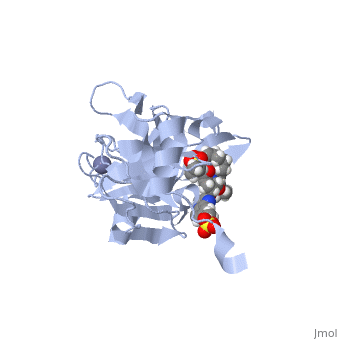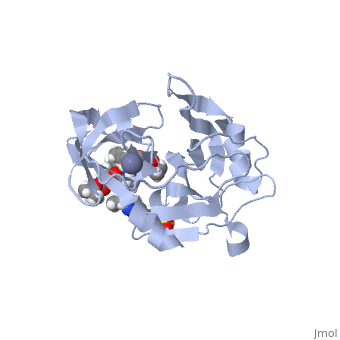
| - | <scene name='74/745998/Grazoprevir/3'>Grazoprevir</scene> (C<sub>38</sub>H<sub>50</sub>N<sub>6</sub>O<sub>9</sub>, molecular weight: 766.911 g/mol) <ref>National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.</ref> stops the hepatitis C virus by inhibiting the NS3/4A protease, which is an enzyme responsible for the cleavage of the viral polyprotein. Cleavage of this polyprotein facilitates viral assembly and maturation of the virus. The NS3/4A protease is a protein complex in which the active site consists of a <scene name='74/746120/Catalytic_triad_of_ns3-4a/1'>catalytic triad</scene> of amino acids S139, H57 and D81, and an activating cofactor which assists in, and is required for the activity of the protease. For more interactive models of the Grazoprevir, click here [http://www.rcsb.org/pdb/explore/explore.do?structureId=5EPY]. Grazoprevir has been proven to be more effective than similar protease inhibitors due to its two-way approach to inhibition. NS3/4A protease cleaves both the viral polyprotein of the Hep C virus and host factors involved in the immune response. Grazoprevir is unique in regard to its P4 cap, and P2 to P4 macrocyclic restriction, which interacts with the catalytic triad of the NS3/4A protease in its own distinct conformation. Due to this conformation, Grazoprevir can easily bind to the catalytic triad because it is already in a highly favorable arrangement, stopping the triads activity.<ref>DOI: 10.1021/acschembio.5b00647</ref>. | + | <scene name='74/745998/Grazoprevir/3'>Grazoprevir</scene> (C<sub>38</sub>H<sub>50</sub>N<sub>6</sub>O<sub>9</sub>, molecular weight: 766.911 g/mol) <ref>National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.</ref> stops the hepatitis C virus by inhibiting the NS3/4A protease, which is an enzyme responsible for the cleavage of the viral polyprotein. Cleavage of this polyprotein facilitates viral assembly and maturation of the virus. The NS3/4A protease is a protein complex in which the active site consists of a <scene name='74/746120/Catalytic_triad_of_ns3-4a/1'>catalytic triad</scene> of amino acids S139, H57 and D81, and an activating cofactor which assists in, and is required for the activity of the protease. For more interactive models of the Grazoprevir, click here [http://www.rcsb.org/pdb/explore/explore.do?structureId=5EPY]. Grazoprevir has been proven to be more effective than similar protease inhibitors due to its two-way approach to inhibition. NS3/4A protease cleaves both the viral polyprotein of the Hep C virus and host factors involved in the immune response. Grazoprevir is unique in regard to its P4 cap, and <scene name='74/746120/P2-p4_macrocylic_ring/1'>P2 to P4 macrocyclic restriction</scene>, which interacts with the catalytic triad of the NS3/4A protease in its own distinct conformation. Due to this conformation, Grazoprevir can easily bind to the catalytic triad because it is already in a highly favorable arrangement, stopping the triads activity.<ref>DOI: 10.1021/acschembio.5b00647</ref>. |
Revision as of 17:41, 5 December 2016
| |||||||||||
References
- ↑ Tan, S.-L. Hepatitis C viruses: genomes and molecular biology; Horizon bioscience: Wymondham, 2006.
- ↑ Chevaliez, S.; Pawlotsky, J. M. Virology of hepatitis C virus infection. Best Pract. Res. Clin. Gastroenterol.2012, 26, 381-389.
- ↑ Tan, S.-L. Hepatitis C viruses: genomes and molecular biology; Horizon bioscience: Wymondham, 2006.
- ↑ Tan, S.-L. Hepatitis C viruses: genomes and molecular biology; Horizon bioscience: Wymondham, 2006.
- ↑ Petrescu, I. O.; Biciusca, V.; Taisescu, C. I.; Alexandru, D. O.; Taisescu, O.; Comanescu, M. V.; Petrescu, F.; Popescu, I. A.; Trasca, D. M.; ForTofoiu, M. C.; Silosi, C. A.; ForTofoiu, M. Histological factors that predict the liver fibrosis in patients with chronic hepatitis C. Rom. J. Morphol. Embryol. 2016, 57, 759-765.
- ↑ 6.0 6.1 Coburn CA, Meinke PT, Chang W, Fandozzi CM, Graham DJ, Hu B, Huang Q, Kargman S, Kozlowski J, Liu R, McCauley JA, Nomeir AA, Soll RM, Vacca JP, Wang D, Wu H, Zhong B, Olsen DB, Ludmerer SW. Discovery of MK-8742: an HCV NS5A inhibitor with broad genotype activity. ChemMedChem. 2013 Dec;8(12):1930-40. doi: 10.1002/cmdc.201300343. Epub 2013 Oct, 14. PMID:24127258 doi:http://dx.doi.org/10.1002/cmdc.201300343
- ↑ Soumana DI, Kurt Yilmaz N, Prachanronarong KL, Aydin C, Ali A, Schiffer CA. Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172. ACS Chem Biol. 2015 Dec 18. PMID:26682473 doi:http://dx.doi.org/10.1021/acschembio.5b00647
- ↑ National Center for Biotechnology Inforamtion. PubChem Compound Database; CID=71661251, https://pubchem.ncbi.nlm.nih.gov/compound/71661251#section=Chemical-and-Physical-Properties
- ↑ http://dx.doi.org/10.1016/j.jhep.2013.03.030
- ↑ Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J Virol. 2011 Jul;85(14):7312-20. doi: 10.1128/JVI.00253-11. Epub 2011 May 18. PMID:21593143 doi:http://dx.doi.org/10.1128/JVI.00253-11
- ↑ National Center for Biotechnology Information. PubChem Compound Database; CID=44603531, https://pubchem.ncbi.nlm.nih.gov/compound/44603531.
- ↑ Soumana DI, Kurt Yilmaz N, Prachanronarong KL, Aydin C, Ali A, Schiffer CA. Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172. ACS Chem Biol. 2015 Dec 18. PMID:26682473 doi:http://dx.doi.org/10.1021/acschembio.5b00647
[1] [2] [3] [4] [5] [6] [7] [8] [9] [10]