1qjg
From Proteopedia
| Line 4: | Line 4: | ||
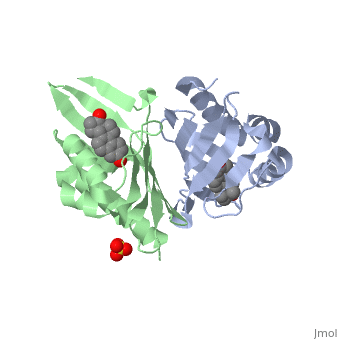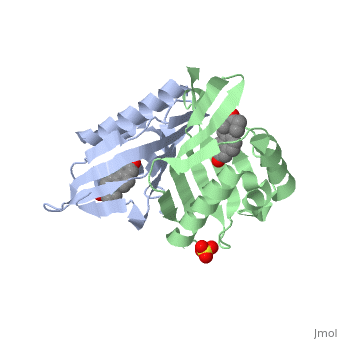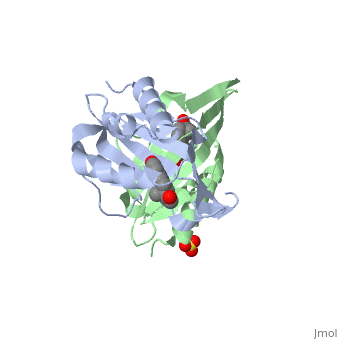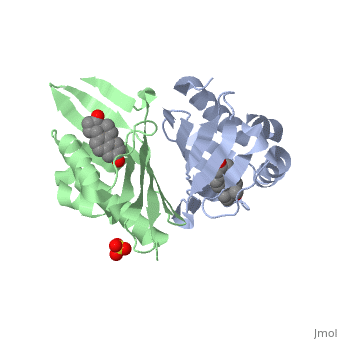
|PDB= 1qjg |SIZE=350|CAPTION= <scene name='initialview01'>1qjg</scene>, resolution 2.3Å | |PDB= 1qjg |SIZE=350|CAPTION= <scene name='initialview01'>1qjg</scene>, resolution 2.3Å | ||
|SITE= | |SITE= | ||
| - | |LIGAND= <scene name='pdbligand= | + | |LIGAND= <scene name='pdbligand=EQU:EQUILENIN'>EQU</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene> |
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1qjg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1qjg OCA], [http://www.ebi.ac.uk/pdbsum/1qjg PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1qjg RCSB]</span> | ||
}} | }} | ||
| Line 24: | Line 27: | ||
[[Category: Cho, H S.]] | [[Category: Cho, H S.]] | ||
[[Category: Oh, B H.]] | [[Category: Oh, B H.]] | ||
| - | [[Category: EQU]] | ||
| - | [[Category: SO4]] | ||
[[Category: isomerase]] | [[Category: isomerase]] | ||
[[Category: ksi]] | [[Category: ksi]] | ||
[[Category: steroid isomeration]] | [[Category: steroid isomeration]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 23:14:31 2008'' |
Revision as of 20:14, 30 March 2008
| |||||||
| , resolution 2.3Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
CRYSTAL STRUCTURE OF DELTA5-3-KETOSTEROID ISOMERASE FROM PSEUDOMONAS TESTOSTERONI IN COMPLEX WITH EQUILENIN
Overview
Delta(5)-3-Ketosteroid isomerase from Pseudomonas testosteroni has been intensively studied as a prototype to understand an enzyme-catalyzed allylic isomerization. Asp(38) (pK(a) approximately 4.7) was identified as the general base abstracting the steroid C4beta proton (pK(a) approximately 12.7) to form a dienolate intermediate. A key and common enigmatic issue involved in the proton abstraction is the question of how the energy required for the unfavorable proton transfer can be provided at the active site of the enzyme and/or how the thermodynamic barrier can be drastically reduced. Answering this question has been hindered by the existence of two differently proposed enzyme reaction mechanisms. The 2.26 A crystal structure of the enzyme in complex with a reaction intermediate analogue equilenin reveals clearly that both the Tyr(14) OH and Asp(99) COOH provide direct hydrogen bonds to the oxyanion of equilenin. The result negates the catalytic dyad mechanism in which Asp(99) donates the hydrogen bond to Tyr(14), which in turn is hydrogen bonded to the steroid. A theoretical calculation also favors the doubly hydrogen-bonded system over the dyad system. Proton nuclear magnetic resonance analyses of several mutant enzymes indicate that the Tyr(14) OH forms a low barrier hydrogen bond with the dienolic oxyanion of the intermediate.
About this Structure
1QJG is a Single protein structure of sequence from Comamonas testosteroni. Full crystallographic information is available from OCA.
Reference
Crystal structure of delta(5)-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization., Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH, J Biol Chem. 1999 Nov 12;274(46):32863-8. PMID:10551849
Page seeded by OCA on Sun Mar 30 23:14:31 2008