This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
P53
From Proteopedia
(Difference between revisions)
| Line 41: | Line 41: | ||
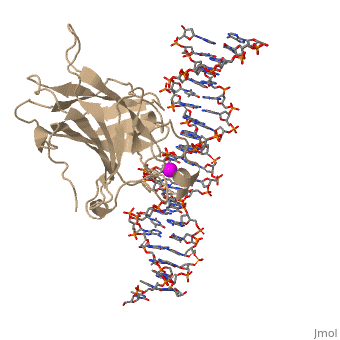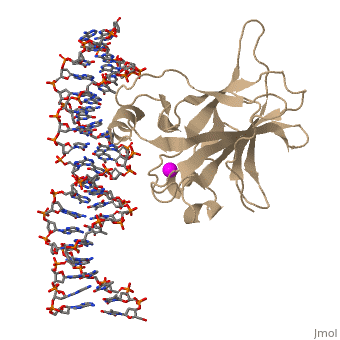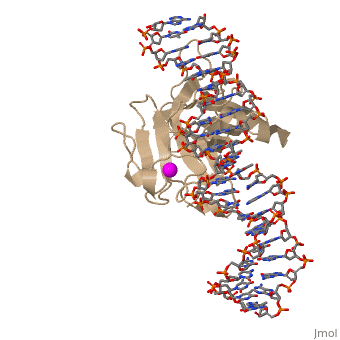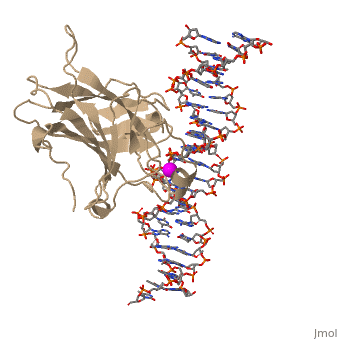
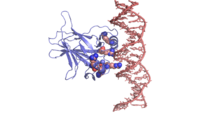
| - | There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues <scene name='26/26327/B_chain_and_dna/ | + | There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues <scene name='26/26327/B_chain_and_dna/12'>C176, H179, C238, and C242</scene> that are located on two loops, respectively. It is conceivable that the zinc plays a role in stabilizing the two loops through |
coordination. The Zn has been represented as a magenta sphere, and R248 in space filling, in the scene at the right. | coordination. The Zn has been represented as a magenta sphere, and R248 in space filling, in the scene at the right. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 16:25, 14 January 2017
| |||||||||||
3D structures of p53 (Updated on 14-January-2017)
Additional Resources
References
- ↑ Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003 Apr;10(4):431-42. PMID:12719720 doi:http://dx.doi.org/10.1038/sj.cdd.4401183
- ↑ Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010 Feb;2(2):a001107. doi:, 10.1101/cshperspect.a001107. PMID:20182618 doi:http://dx.doi.org/10.1101/cshperspect.a001107
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mary Ball, Alexander Berchansky, David Canner