User:Morgane Crausaz/Sandbox
From Proteopedia
| Line 15: | Line 15: | ||
The crystal structure was performed by 1,83 Angstrom spacing. The researchers used a protein secreted from HEK293S GnTI- cells. those cells come from embryonic kidney of human and do not express N-acetylglucosaminyltransferase I. This line of cells is used to overexpress a wide variety of mammalian membrane proteins. | The crystal structure was performed by 1,83 Angstrom spacing. The researchers used a protein secreted from HEK293S GnTI- cells. those cells come from embryonic kidney of human and do not express N-acetylglucosaminyltransferase I. This line of cells is used to overexpress a wide variety of mammalian membrane proteins. | ||
| - | The lysosomal phospholipase A2 is composed of 380 amino acids. The protein contains both alpha helix and beta strand therefore, belongs to the alpha/beta hydrolase superfamily. The protein is divided in three domains. First, the membrane binding domain from Leucine 18 to Proline 81, contains six beta strand and two alpha helixes. The cap domain from Glycine 198 to Threonine 288 then, from Glycine 301 to Glycine 324 presents five alpha helix and four beta strand. Finally, the hydrolase domain from Alanine 1 to Glutamine 17, from Glycine 82 to Glycine 197, from Methionine 289 to Threonine 300, then, from Aspartate 325 to the end of the protein. | + | The lysosomal phospholipase A2 is composed of 380 amino acids. The protein contains both alpha helix and beta strand therefore, belongs to the '''alpha/beta hydrolase superfamily'''. [[Image:Example.jpg]] |
| - | The lysosomal phospholipase A2 has a catalytic triad in the alpha/beta hydrolase domain at conserved topological positions: Serine 165, Aspartate 327 and Histidine 359. Some of particularity of the protein are observed in the protein structure: there is a disulfide bond between Cysteine 32 and Cysteine 56; a lid loop from Aspartate 211 to Proline 220 and this characteristic loop is a conserved element in different species. This loop undergoes a conformational change upon contact with its substrate to allow the substrate to access the active site of the protein. | + | |
| + | The protein is divided in '''three domains'''. First, the <Structure load='membrane binding domain' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> from Leucine 18 to Proline 81, contains six beta strand and two alpha helixes. The <Structure load='cap domain' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> from Glycine 198 to Threonine 288 then, from Glycine 301 to Glycine 324 presents five alpha helix and four beta strand. Finally, the <Structure load='hydrolase domain' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> from Alanine 1 to Glutamine 17, from Glycine 82 to Glycine 197, from Methionine 289 to Threonine 300, then, from Aspartate 325 to the end of the protein. | ||
| + | |||
| + | The lysosomal phospholipase A2 has a <Structure load='catalytic triad' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> in the alpha/beta hydrolase domain at conserved topological positions: Serine 165, Aspartate 327 and Histidine 359. Some of particularity of the protein are observed in the protein structure: there is a <Structure load='disulfide bond' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> between Cysteine 32 and Cysteine 56; a <Structure load='lid loop' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> from Aspartate 211 to Proline 220 and this characteristic loop is a conserved element in different species. This loop undergoes a conformational change upon contact with its substrate to allow the substrate to access the active site of the protein. | ||
==Function== | ==Function== | ||
Revision as of 12:28, 24 January 2017
|
This is a default text for your page Lysosomal phospholipase A2. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Contents |
Introduction
The lysosome is a membrane-bound organelle which is present in animal cells. Those are acidic vesicles and contain more than fifty digestive enzymes such as proteases, nucleases, glycosidases, sulfatases, lipases, phosphatases phospholipases and esterases. The lumen’s pH of 4,5 is optimal for the hydrolytic enzymes. Indeed, acidic pH is important for the degradation of intracellular and extracellular compounds.
Particularly, there are enzymes able to cleave phospholipids on a specific site: those enzymes are named: phospholipases. These phospholipases are classified into four types, phospholipase A1, A2, C and D depending on the specific cleavage site of the substrate. Phospholipase A2 cleave the acyl-ester bonds of sn-2 position of glycerophospholipids and they produce free fatty acids.
Substrate specificity depending on the phospholipase family :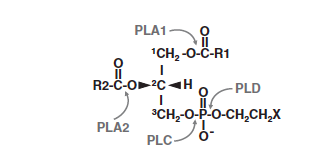
Structure
The crystal structure was performed by 1,83 Angstrom spacing. The researchers used a protein secreted from HEK293S GnTI- cells. those cells come from embryonic kidney of human and do not express N-acetylglucosaminyltransferase I. This line of cells is used to overexpress a wide variety of mammalian membrane proteins.
The lysosomal phospholipase A2 is composed of 380 amino acids. The protein contains both alpha helix and beta strand therefore, belongs to the alpha/beta hydrolase superfamily. 
|
|
|
|
|
|
Function
The lysosomal phospholipase A2 has a global basic electrostatic surface that is complementary of the acidic inner membrane of the lysosomal membrane. The structure shows a hydrophobic surface including Tyrosine 30, Leucine 31, Leucine 50 and Valine 52 on the membrane-binding domain which binds to the lipid bilayers.
After docking on membrane, a phospholipid, substrate of the enzyme, enters into the active site. The catalytic triad is located such a way to cleave the acyl group of the phospholipids. The serine is able to act as a nucleophile cleave the acyl-ester bonds. The histidine of the catalytic triad is placed to protonate the lysophospholipid after the cleavage.
Ligands
Disease's treatment
Implication of the lysosomal phospholipase A2 in the detoxification :
Lipid oxidation products and in particular oxidized phospholipids (OxPL) are increasingly recognized as inducers of chronic inflammation characteristic of atherosclerosis. Atherosclerosis is a chronic inflammatory disease characterized by accumulation of monocytes and T-cells due to lipid abnormalities. Increased levels of phospholipids’ oxidation products have been detected in different organs and pathological states, including atherosclerotic vessels. They can integrate the lipid membranes of cells and lipoproteins, act as ligands and may cause local membrane disruption. Indeed, they stimulate production of chemokines and adhesion of monocytes to endothelial cells. Truncated oxidized-glycerophospholipids (ox-PLs) are bioactive lipids resulting from oxidative stress. They are generated by the oxidation of polyunsaturated fatty acid residues, which are usually present in the phospholipids at the sn-2 position (cleave position of lysosomal phospholipase A2). Since the ox-PLs are transferred to lysosomes, the lysosomal phospholipase A2 plays an important role in the degradation of them. In fact, lysosomal phospholipase A2 preferentially hydrolyzes truncated ox-PCs compared to non-oxidized phospholipids with two long acyl chains (like DOPC) under acidic conditions
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
