User:Blandine Velut/Sandbox
From Proteopedia
| Line 12: | Line 12: | ||
== Structure == | == Structure == | ||
| - | GMP synthetase is a homodimer enzyme, in which each monomer is composed of 693 amino acids and weights 76,2 kDa | + | GMP synthetase is a homodimer enzyme, in which each monomer is composed of 693 amino acids and weights 76,2 kDa <ref>http://www.uniprot.org/uniprot/P49915</ref>. Each monomer is composed of <scene name='75/750228/2_domains/2'>two catalytic domains</scene> , encoded by a single gene: a N-terminal glutaminase domain (GATase domain), and a C-terminal synthetase domain. |
[[Image:GMPs.jpg|thumb|center|750px|Secondary structure of GMP synthetase]] | [[Image:GMPs.jpg|thumb|center|750px|Secondary structure of GMP synthetase]] | ||
Revision as of 19:50, 26 January 2017
2vxo
HUMAN GMP SYNTHETASE
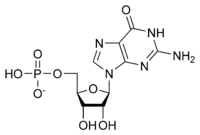
An ample supply of nucleotides is essential for many life processes, including cell maturation, cell division and transmission of the genetic information. Indeed, nucleotides are the activated precursors of nucleic acids, but they also are major energy carriers, and precursors for the synthesis of nucleotide cofactors. Among these molecules is the guanosine monophosphate (GMP), also known as 5'-guanidylic acid or guanylic acid, a nucleotide that is used as a monomer in RNA. Like other nucleotides, GMP can be synthesized by 2 main pathways : de novo pathway and salvage pathway. De novo synthesis of nucleotide involves several enzymatic reaction and enzymes. Here, we will focus on the final step of the process, which is catalyzed by a glutamine amidotransferase called GMP synthetase (GMPS; E.C. 6.3.5.2). This enzyme belongs to the family of ligases, and catalyzes the conversion of xanthine monophosphate (XMP) to GMP in the presence of glutamine and ATP. [1]
| |||||||||||
References
- ↑ Oliver JC, Linger RS, Chittur SV, Davisson VJ. Substrate activation and conformational dynamics of guanosine 5'-monophosphate synthetase. Biochemistry. 2013 Aug 6;52(31):5225-35. doi: 10.1021/bi3017075. Epub 2013 Jul, 23. PMID:23841499 doi:http://dx.doi.org/10.1021/bi3017075
- ↑ http://www.uniprot.org/uniprot/P49915