User:Blandine Velut/Sandbox
From Proteopedia
| Line 11: | Line 11: | ||
== Structure == | == Structure == | ||
| - | GMP synthetase is a homodimer enzyme, in which each monomer is composed of 693 amino acids and weights 76,2 kDa <ref>http://www.uniprot.org/uniprot/P49915</ref>. Each monomer is composed of <scene name='75/750228/2_domains/2'>two catalytic domains</scene> , encoded by a single gene: a N-terminal glutaminase domain (GATase domain), and a C-terminal synthetase domain. | + | GMP synthetase is a homodimer enzyme, in which each monomer is composed of 693 amino acids and weights 76,2 kDa <ref>http://www.uniprot.org/uniprot/P49915</ref>. Each monomer is composed of <scene name='75/750228/2_domains/2'>two catalytic domains</scene> , encoded by a single gene: a N-terminal glutaminase domain (GATase domain), and a C-terminal synthetase domain. |
[[Image:GMPs.jpg|thumb|center|750px|Secondary structure of GMP synthetase]] | [[Image:GMPs.jpg|thumb|center|750px|Secondary structure of GMP synthetase]] | ||
| Line 18: | Line 18: | ||
'''GTase domain''' | '''GTase domain''' | ||
| - | The GATase domain, stretched from residue 27 to residue 216, is composed of a single structural domain. It is constituted of a central β-sheet surrounded by several α-helices. It contains <scene name='75/750228/Triad/1'>the catalytic triad</scene> composed of residues Cys104, His190, and Glu192. | + | The GATase domain, stretched from residue 27 to residue 216, is composed of a single structural domain. It is constituted of a central β-sheet surrounded by several α-helices. It contains <scene name='75/750228/Triad/1'>the catalytic triad</scene> composed of residues Cys104, His190, and Glu192. |
| Line 33: | Line 33: | ||
<scene name='75/750228/Active_site/1'>The active site</scene> is located between the ATPPase sub-domain and the D2 sub-domain. When XMP is bound to it, it is allostericly regulated and covered by a lid motif (residues 368-408) closing in over the active site. Thus, the XMP is wedged between a Pro-rich region (residues 438–441) and a loop (residues 383–385). | <scene name='75/750228/Active_site/1'>The active site</scene> is located between the ATPPase sub-domain and the D2 sub-domain. When XMP is bound to it, it is allostericly regulated and covered by a lid motif (residues 368-408) closing in over the active site. Thus, the XMP is wedged between a Pro-rich region (residues 438–441) and a loop (residues 383–385). | ||
| - | The synthetase domain binds several cofactors. Indeed, <scene name='75/750228/Sulfate/1'>three sulphate ions</scene> are bound to the ATPPase and D2 sub-domains. There are also Mg2+ and ATP which can bind. | + | The synthetase domain binds several cofactors. Indeed, <scene name='75/750228/Sulfate/1'>three sulphate ions</scene> are bound to the ATPPase and D2 sub-domains. There are also Mg2+ and ATP which can bind. |
There are 2 isoforms produced by alternative splicing: the isoform 1, which was just described, and the isoform 2. The sequence of this isoform differs from the first isoform by the missing sequence from residue 10 to residue 108. Thus, this isoform is only composed of 594 amino acids and weights 65,9 kDa <ref>http://www.uniprot.org/uniprot/P49915</ref>. | There are 2 isoforms produced by alternative splicing: the isoform 1, which was just described, and the isoform 2. The sequence of this isoform differs from the first isoform by the missing sequence from residue 10 to residue 108. Thus, this isoform is only composed of 594 amino acids and weights 65,9 kDa <ref>http://www.uniprot.org/uniprot/P49915</ref>. | ||
| Line 43: | Line 43: | ||
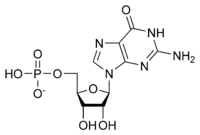
[[Image:GMP.png|thumb|right|500px|GMP]] | [[Image:GMP.png|thumb|right|500px|GMP]] | ||
| - | GMP synthetase is a cytosolic enzyme belonging to the glutamine amidotransferases family. These amidotransferases catalyse the amination of a wide range of molecules using the amide nitrogen of the side chain of glutamine. GMP synthetase is one of the three glutamine amidotransferases that plays a role in the ''de novo'' purine biosynthesis <ref>PMID: 23841499</ref>. Indeed, thanks to its bifunctional two domains, GMP synthetase catalyses the final step in the ''de novo'' synthesis of GMP from XMP in the presence of other cofactors including ATP, glutamine and water. | + | GMP synthetase is a cytosolic enzyme belonging to the glutamine amidotransferases family. These amidotransferases catalyse the amination of a wide range of molecules using the amide nitrogen of the side chain of glutamine. GMP synthetase is one of the three glutamine amidotransferases that plays a role in the ''de novo'' purine biosynthesis <ref>PMID: 23841499</ref>. Indeed, thanks to its bifunctional two domains, GMP synthetase catalyses the final step in the ''de novo'' synthesis of GMP from XMP in the presence of other cofactors including ATP, glutamine and water. The global reaction is summarized below: |
ATP + XMP + L-glutamine + H2O --> AMP + diphosphate + GMP + L-glutamate. | ATP + XMP + L-glutamine + H2O --> AMP + diphosphate + GMP + L-glutamate. | ||
| Line 51: | Line 51: | ||
<br />2) ATP + XMP + NH3 --> AMP + pyrophosphate + GMP | <br />2) ATP + XMP + NH3 --> AMP + pyrophosphate + GMP | ||
| - | First, the glutaminase domain generates ammonia from glutamine-hydrolysis when L-glutamine binds to the catalytic triad. Then, an activation step prepares the XMP for amination. Indeed, GPM synthetase activates its XMP substrate by adenylylation on the xanthine C2 oxygen, which can then be primed for attack by a nitrogen nucleophile. In order to perform the second reaction, the glutamine-derived ammonia needs to be transferred to the <scene name='75/750228/Xmp/1'>XMP</scene>. This one is located in the active site of the synthetase domain, situated no far away from the catalytic triad. The ammonia translocation is enabled by a channel between the two active sites. This channel is formed thanks to a conformational change of the catalytic triad, further to its production. Thus, the activated XMP is aminated to produce GMP. Then, the GMP is released and will be used as a monomer in RNA. | + | First, the glutaminase domain generates ammonia from glutamine-hydrolysis when L-glutamine binds to the catalytic triad. Then, an activation step prepares the XMP for amination. Indeed, GPM synthetase activates its XMP substrate by adenylylation on the xanthine C2 oxygen, which can then be primed for attack by a nitrogen nucleophile. In order to perform the second reaction, the glutamine-derived ammonia needs to be transferred to the <scene name='75/750228/Xmp/1'>XMP</scene>. This one is located in the active site of the synthetase domain, situated no far away from the catalytic triad. The ammonia translocation is enabled by a channel between the two active sites. This channel is formed thanks to a conformational change of the catalytic triad, further to its production. Thus, the activated XMP is aminated to produce GMP. Then, the GMP is released and will be used as a monomer in RNA. |
| Line 58: | Line 58: | ||
| - | The presence of free Mg2+ is essential for activation of the GMP synthetase and a complex between ATP and Mg2+ can be formed but MgATP2− alone is not sufficient for catalysis. Moreover, the total chelation of free Mg2+ by ATP results of inactivation of the enzyme. | + | The presence of free Mg2+ is essential for activation of the GMP synthetase and a complex between ATP and Mg2+ can be formed but MgATP2− alone is not sufficient for catalysis. Moreover, the total chelation of free Mg2+ by ATP results of inactivation of the enzyme. <ref>10.1074/jbc.270.13.7347</ref> |
'''Inhibitors''' | '''Inhibitors''' | ||
Revision as of 21:12, 26 January 2017
2vxo
HUMAN GMP SYNTHETASE
An ample supply of nucleotides is essential for many life processes, including cell maturation, cell division and transmission of the genetic information. Indeed, nucleotides are the activated precursors of nucleic acids, but they also are major energy carriers, and precursors for the synthesis of nucleotide cofactors. Among these molecules is the guanosine monophosphate (GMP), also known as 5'-guanidylic acid or guanylic acid, a nucleotide that is used as a monomer in RNA. Like other nucleotides, GMP can be synthesized by 2 main pathways : de novo pathway and salvage pathway. De novo synthesis of nucleotide involves several enzymatic reaction and enzymes. Here, we will focus on the final step of the process, which is catalyzed by a glutamine amidotransferase called GMP synthetase (GMPS; E.C. 6.3.5.2). This enzyme belongs to the family of ligases, and catalyzes the conversion of xanthine monophosphate (XMP) to GMP in the presence of glutamine and ATP. [1]
| |||||||||||
References
- ↑ Welin M, Lehtio L, Johansson A, Flodin S, Nyman T, Tresaugues L, Hammarstrom M, Graslund S, Nordlund P. Substrate Specificity and Oligomerization of Human GMP Synthetase. J Mol Biol. 2013 Jun 28. pii: S0022-2836(13)00427-0. doi:, 10.1016/j.jmb.2013.06.032. PMID:23816837 doi:10.1016/j.jmb.2013.06.032
- ↑ http://www.uniprot.org/uniprot/P49915
- ↑ http://www.uniprot.org/uniprot/P49915
- ↑ Oliver JC, Linger RS, Chittur SV, Davisson VJ. Substrate activation and conformational dynamics of guanosine 5'-monophosphate synthetase. Biochemistry. 2013 Aug 6;52(31):5225-35. doi: 10.1021/bi3017075. Epub 2013 Jul, 23. PMID:23841499 doi:http://dx.doi.org/10.1021/bi3017075
- ↑ 10.1074/jbc.270.13.7347
- ↑ Lui MS, Kizaki H, Weber G. Biochemical pharmacology of acivicin in rat hepatoma cells. Biochem Pharmacol. 1982 Nov 1;31(21):3469-73. PMID:7150366
- ↑ Oliver JC, Linger RS, Chittur SV, Davisson VJ. Substrate activation and conformational dynamics of guanosine 5'-monophosphate synthetase. Biochemistry. 2013 Aug 6;52(31):5225-35. doi: 10.1021/bi3017075. Epub 2013 Jul, 23. PMID:23841499 doi:http://dx.doi.org/10.1021/bi3017075
- ↑ Pegram LD, Megonigal MD, Lange BJ, Nowell PC, Rowley JD, Rappaport EF, Felix CA. t(3;11) translocation in treatment-related acute myeloid leukemia fuses MLL with the GMPS (GUANOSINE 5' MONOPHOSPHATE SYNTHETASE) gene. Blood. 2000 Dec 15;96(13):4360-2. PMID:11110714
- ↑ Rodriguez-Suarez R, Xu D, Veillette K, Davison J, Sillaots S, Kauffman S, Hu W, Bowman J, Martel N, Trosok S, Wang H, Zhang L, Huang LY, Li Y, Rahkhoodaee F, Ransom T, Gauvin D, Douglas C, Youngman P, Becker J, Jiang B, Roemer T. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem Biol. 2007 Oct;14(10):1163-75. PMID:17961828 doi:http://dx.doi.org/10.1016/j.chembiol.2007.09.009
- ↑ Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011 Oct 7;44(1):147-59. PMID:21981925 doi:10.1016/j.molcel.2011.06.034