We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Estelle Metzger/Sandbox
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
Roco proteins are serine/threonine specific kinases. This family consists of multidomain Ras-GTPases. Roco4 is 193 kDa and is identified as a key protein for proper stalk cell formation. Between the ''Dictyostelium'' Roco genes and LRRK genes, there are many structural similarities, which are due to independant acquisitions of distantly related protein kinase domain. | Roco proteins are serine/threonine specific kinases. This family consists of multidomain Ras-GTPases. Roco4 is 193 kDa and is identified as a key protein for proper stalk cell formation. Between the ''Dictyostelium'' Roco genes and LRRK genes, there are many structural similarities, which are due to independant acquisitions of distantly related protein kinase domain. | ||
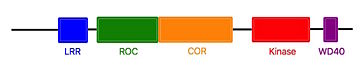
| - | The characteristics of roco protein family, are a conserved core, consisting of a Ras-like GTPase domain called ROC (Ras of Complex proteins) and a COR domain (C-terminal of ROC), a C-terminal kinase domain and several N-terminal leucine rich repeats (LRR). Roco4 possesses one more domain : a C-terminal WD40 repeats.<ref name="Bernd2"/> | + | The characteristics of roco protein family, are a conserved core, consisting of a Ras-like GTPase domain called ROC (Ras of Complex proteins) and a COR domain (C-terminal of ROC), a C-terminal <scene name='75/751216/Roc/2'>kinase domain</scene> and several N-terminal leucine rich repeats (LRR). Roco4 possesses one more domain : a C-terminal WD40 repeats.<ref name="Bernd2"/> |
[[Image:Roco4.jpg|thumb| Linear structure of Roco4 <ref name="Bernd2">doi: 10.3389/fnmol.2014.00032</ref>|center|upright=2,5]] | [[Image:Roco4.jpg|thumb| Linear structure of Roco4 <ref name="Bernd2">doi: 10.3389/fnmol.2014.00032</ref>|center|upright=2,5]] | ||
| Line 16: | Line 16: | ||
The COR and the ROC domains forms an inseperable tandem, a 300-400 long stretch of amino acids with no significant homology to other described domains. <ref name="Bernd2"/> | The COR and the ROC domains forms an inseperable tandem, a 300-400 long stretch of amino acids with no significant homology to other described domains. <ref name="Bernd2"/> | ||
| - | The Roco4 kinase structure consists of a canonical, two-lobed kinase structure, with an adenine nucleotide bound in the conventional | + | The Roco4 kinase structure consists of a canonical, two-lobed kinase structure, with an adenine nucleotide bound in the conventional <scene name='75/751216/Atp_binding_pocket/1'>ATP-binding pocket</scene>. It contains the conserved alphaC-helix and an anti-parallel beta sheets in the smaller N-terminal lobe. Other Alpha-helices and the activation loop with the conserved N-terminal DFG motif are localized in the bigger C-terminal lobe.<ref name="Bernd2"/> |
The activation loop and alphaC-helix together form the catalytic site of the kinase, an ATP binding site formed by a cleft between the two lobes. | The activation loop and alphaC-helix together form the catalytic site of the kinase, an ATP binding site formed by a cleft between the two lobes. | ||
For catalysis, the formation of a polar contact is essential. This polar contact takes place between Roco4 Lys1055 from the beta3-strand and the Glu1078 from the alphaC-helix. The amino acids Asp makes contact with all three ATP phosphates either directly or via coordination of a magnesium ion. Moreover, the amino acid Phe makes hydrophobic contacts to the alphaC-helix and the HxD motif, and leads for the correct positioning of the DFG motif. <ref name="Bernd2"/> | For catalysis, the formation of a polar contact is essential. This polar contact takes place between Roco4 Lys1055 from the beta3-strand and the Glu1078 from the alphaC-helix. The amino acids Asp makes contact with all three ATP phosphates either directly or via coordination of a magnesium ion. Moreover, the amino acid Phe makes hydrophobic contacts to the alphaC-helix and the HxD motif, and leads for the correct positioning of the DFG motif. <ref name="Bernd2"/> | ||
Revision as of 22:41, 26 January 2017
Humanized Roco4 bound to LRRK2-IN-1
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Gilsbach BK, Messias AC, Ito G, Sattler M, Alessi DR, Wittinghofer A, Kortholt A. Structural Characterization of LRRK2 Inhibitors. J Med Chem. 2015 May 1. PMID:25897865 doi:http://dx.doi.org/10.1021/jm5018779
- ↑ 2.0 2.1 2.2 2.3 2.4 Gilsbach BK, Kortholt A. Structural biology of the LRRK2 GTPase and kinase domains: implications for regulation. Front Mol Neurosci. 2014 May 5;7:32. doi: 10.3389/fnmol.2014.00032. eCollection, 2014. PMID:24847205 doi:http://dx.doi.org/10.3389/fnmol.2014.00032
- ↑ 3.0 3.1 doi: https://dx.doi.org/10.1016/S0092-8674(02)00741-9
- ↑ 4.0 4.1 Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011 Feb;36(2):65-77. doi: 10.1016/j.tibs.2010.09.006. Epub, 2010 Oct 23. PMID:20971646 doi:10.1016/j.tibs.2010.09.006