We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Estelle Metzger/Sandbox
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
Some studies to find a treatment for the Parkinson's disease are focused on LRRK2. Indeed, mutations in LRRK2, which increases its kinase activity, are found in case of Parkinson’s disease. Thus, a kinase inhibitor for LRRK2 would be an interesting thetapeutic target. | Some studies to find a treatment for the Parkinson's disease are focused on LRRK2. Indeed, mutations in LRRK2, which increases its kinase activity, are found in case of Parkinson’s disease. Thus, a kinase inhibitor for LRRK2 would be an interesting thetapeutic target. | ||
| - | Thanks to the similarity between LRRK2 and Roco4 from the ''Dictyostelium'', Roco4 is used in studies with a view to finding that inhibitor. One of the candidates to inhibit this activity is <scene name='75/751216/Lrrk2- | + | Thanks to the similarity between LRRK2 and Roco4 from the ''Dictyostelium'', Roco4 is used in studies with a view to finding that inhibitor. One of the candidates to inhibit this activity is <scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene>.<ref name="Bernd">doi: 10.1021/jm5018779</ref> |
| Line 9: | Line 9: | ||
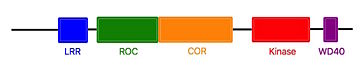
Roco proteins are serine/threonine specific kinases. This family consists of multidomain Ras-GTPases. Roco4 is 193 kDa and is identified as a key protein for proper stalk cell formation. Between the ''Dictyostelium'' Roco genes and LRRK genes, there are many structural similarities, which are due to independant acquisitions of distantly related protein kinase domain. | Roco proteins are serine/threonine specific kinases. This family consists of multidomain Ras-GTPases. Roco4 is 193 kDa and is identified as a key protein for proper stalk cell formation. Between the ''Dictyostelium'' Roco genes and LRRK genes, there are many structural similarities, which are due to independant acquisitions of distantly related protein kinase domain. | ||
| - | The characteristics of roco protein family, are a conserved core, consisting of a Ras-like GTPase domain called ROC (Ras of Complex proteins) and a COR domain (C-terminal of ROC), a C-terminal <scene name='75/751216/ | + | The characteristics of roco protein family, are a conserved core, consisting of a Ras-like GTPase domain called ROC (Ras of Complex proteins) and a COR domain (C-terminal of ROC), a C-terminal <scene name='75/751216/Kinase_domain/1'>kinase domain</scene> and several N-terminal leucine rich repeats (LRR). Roco4 possesses one more domain : a C-terminal WD40 repeats.<ref name="Bernd2"/> |
[[Image:Roco4.jpg|thumb| Linear structure of Roco4 <ref name="Bernd2">doi: 10.3389/fnmol.2014.00032</ref>|center|upright=2,5]] | [[Image:Roco4.jpg|thumb| Linear structure of Roco4 <ref name="Bernd2">doi: 10.3389/fnmol.2014.00032</ref>|center|upright=2,5]] | ||
| Line 30: | Line 30: | ||
== LRRK2-IN-1 == | == LRRK2-IN-1 == | ||
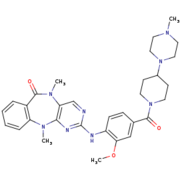
[[Image:4K4-270.png|thumb|LRRK2-IN-1 structure (4K4 on PDB website)|upright=1,5]] | [[Image:4K4-270.png|thumb|LRRK2-IN-1 structure (4K4 on PDB website)|upright=1,5]] | ||
| - | <scene name='75/751216/Lrrk2- | + | <scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene> is the first identified LRRK2-specific inhibitor, which is now a common tool compound for the LRRK2 research community. LRRK2-IN-1 has a 2-amino-5,11- dimethyl-5H-benzo[e]pyrimido[5,4-b][1,4]diazepine-6(11H)-one scaffold. |
The function is of LRRK2-In-1 is to dephosphorylate LRRK2 residues Ser910 and Ser935 in the kidney, but not in the brain. This compound is not capable of crossing the blood-brain barrier. | The function is of LRRK2-In-1 is to dephosphorylate LRRK2 residues Ser910 and Ser935 in the kidney, but not in the brain. This compound is not capable of crossing the blood-brain barrier. | ||
The structure of LRRK2-In-1 does not stabilize the active conformation. Indeed, the activation loop is poorly resolved indicating that it is flexible. Moreover, it presents a closure of the glycine-rich loop in the inhibitor structure.<ref name="Bernd"/> | The structure of LRRK2-In-1 does not stabilize the active conformation. Indeed, the activation loop is poorly resolved indicating that it is flexible. Moreover, it presents a closure of the glycine-rich loop in the inhibitor structure.<ref name="Bernd"/> | ||
| Line 42: | Line 42: | ||
LRRK2-IN-1 makes 2 hydrogen bounds and 24 vander Walls contacts with Roco4. The first hydrogen bound is formed between the backbone carbonyl of Val1055 and the N24 of the LRRK2-IN-1 with a distance of 2.8 Å. The second is formed between the Nz of Lys1055 and O40 of LRRK2-IN-1 with a | LRRK2-IN-1 makes 2 hydrogen bounds and 24 vander Walls contacts with Roco4. The first hydrogen bound is formed between the backbone carbonyl of Val1055 and the N24 of the LRRK2-IN-1 with a distance of 2.8 Å. The second is formed between the Nz of Lys1055 and O40 of LRRK2-IN-1 with a | ||
| - | distance of 3.2 Å. This interaction takes place in the ATP binding pocket<ref name="Bernd"/>. | + | distance of 3.2 Å. This interaction takes place in the <scene name='75/751216/Atp_binding_pocket/1'>ATP-binding pocket</scene><ref name="Bernd"/>. |
LRRK2-IN-1 is predicted to be a type 1 inhibitor. That mean that it should only bind the active form of its target and stabilize it. However, a study showed that it can also bind the inactive form of Roco4<ref name="Bernd"/>. | LRRK2-IN-1 is predicted to be a type 1 inhibitor. That mean that it should only bind the active form of its target and stabilize it. However, a study showed that it can also bind the inactive form of Roco4<ref name="Bernd"/>. | ||
Revision as of 22:52, 26 January 2017
Humanized Roco4 bound to LRRK2-IN-1
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Gilsbach BK, Messias AC, Ito G, Sattler M, Alessi DR, Wittinghofer A, Kortholt A. Structural Characterization of LRRK2 Inhibitors. J Med Chem. 2015 May 1. PMID:25897865 doi:http://dx.doi.org/10.1021/jm5018779
- ↑ 2.0 2.1 2.2 2.3 2.4 Gilsbach BK, Kortholt A. Structural biology of the LRRK2 GTPase and kinase domains: implications for regulation. Front Mol Neurosci. 2014 May 5;7:32. doi: 10.3389/fnmol.2014.00032. eCollection, 2014. PMID:24847205 doi:http://dx.doi.org/10.3389/fnmol.2014.00032
- ↑ 3.0 3.1 doi: https://dx.doi.org/10.1016/S0092-8674(02)00741-9
- ↑ 4.0 4.1 Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011 Feb;36(2):65-77. doi: 10.1016/j.tibs.2010.09.006. Epub, 2010 Oct 23. PMID:20971646 doi:10.1016/j.tibs.2010.09.006