We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Estelle Metzger/Sandbox
From Proteopedia
(Difference between revisions)
| Line 54: | Line 54: | ||
The use of roco4, permited to learn that the G2019S mutation is the results of an additional hydrogen bound between Ser2019 (Ser1179 in Roco4) and Gln1918 (Arg1077 in Roco4).<ref name="Bernd"/><ref name="Mills"/> | The use of roco4, permited to learn that the G2019S mutation is the results of an additional hydrogen bound between Ser2019 (Ser1179 in Roco4) and Gln1918 (Arg1077 in Roco4).<ref name="Bernd"/><ref name="Mills"/> | ||
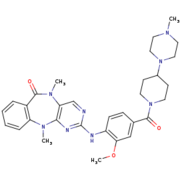
| - | However, <scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene>, like many LRRK2 inhibitors, presents a lack of selectivity or difficulties to pass the blood-brain barrier. Kinase inhibitors can lead also to kidney and lung abnormality. Without working on these points,<scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene> couldn’t be used as a treatment for the Parkinson’s disease. | + | However, <scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene>, like many LRRK2 inhibitors, presents a lack of selectivity or difficulties to pass the blood-brain barrier. Kinase inhibitors can lead also to kidney and lung abnormality. Without working on these points,<scene name='75/751216/Lrrk2-in-1/3'>LRRK2-IN-1</scene> couldn’t be used as a treatment for the Parkinson’s disease. In the main time, they are used as tools for characterization of the function and activation mechanism of LRRK2 in order to find new therapeutic target. <ref name="Bernd"/> |
| - | In the main time, they are used as tools for characterization of the function and activation mechanism of LRRK2 in order to find new therapeutic target. <ref name="Bernd"/> | + | |
Revision as of 09:12, 27 January 2017
Humanized Roco4 bound to LRRK2-IN-1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Gilsbach BK, Ho FY, Vetter IR, van Haastert PJ, Wittinghofer A, Kortholt A. Roco kinase structures give insights into the mechanism of Parkinson disease-related leucine-rich-repeat kinase 2 mutations. Proc Natl Acad Sci U S A. 2012 Jun 11. PMID:22689969 doi:10.1073/pnas.1203223109
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Gilsbach BK, Messias AC, Ito G, Sattler M, Alessi DR, Wittinghofer A, Kortholt A. Structural Characterization of LRRK2 Inhibitors. J Med Chem. 2015 May 1. PMID:25897865 doi:http://dx.doi.org/10.1021/jm5018779
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 van Egmond WN, van Haastert PJ. Characterization of the Roco protein family in Dictyostelium discoideum. Eukaryot Cell. 2010 May;9(5):751-61. doi: 10.1128/EC.00366-09. Epub 2010 Mar 26. PMID:20348387 doi:http://dx.doi.org/10.1128/EC.00366-09
- ↑ 4.0 4.1 4.2 4.3 4.4 Gilsbach BK, Kortholt A. Structural biology of the LRRK2 GTPase and kinase domains: implications for regulation. Front Mol Neurosci. 2014 May 5;7:32. doi: 10.3389/fnmol.2014.00032. eCollection, 2014. PMID:24847205 doi:http://dx.doi.org/10.3389/fnmol.2014.00032
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 Mills RD, Mulhern TD, Liu F, Culvenor JG, Cheng HC. Prediction of the repeat domain structures and impact of parkinsonism-associated variations on structure and function of all functional domains of leucine-rich repeat kinase 2 (LRRK2). Hum Mutat. 2014 Apr;35(4):395-412. doi: 10.1002/humu.22515. Epub 2014 Feb 24. PMID:24470158 doi:http://dx.doi.org/10.1002/humu.22515
- ↑ 6.0 6.1 Liu Z, West AB. The dual enzyme LRRK2 hydrolyzes GTP in both its GTPase and kinase domains in vitro. Biochim Biophys Acta. 2016 Dec 8;1865(3):274-280. doi:, 10.1016/j.bbapap.2016.12.001. PMID:27939437 doi:http://dx.doi.org/10.1016/j.bbapap.2016.12.001
- ↑ 7.0 7.1 doi: https://dx.doi.org/10.1016/S0092-8674(02)00741-9
- ↑ 8.0 8.1 Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011 Feb;36(2):65-77. doi: 10.1016/j.tibs.2010.09.006. Epub, 2010 Oct 23. PMID:20971646 doi:10.1016/j.tibs.2010.09.006
- ↑ [1], Retrieved on January 27th 2017.
- ↑ 10.0 10.1 UniProtKB - Q5S007 (LRRK2_HUMAN), Retrieved on January 27th 2017.